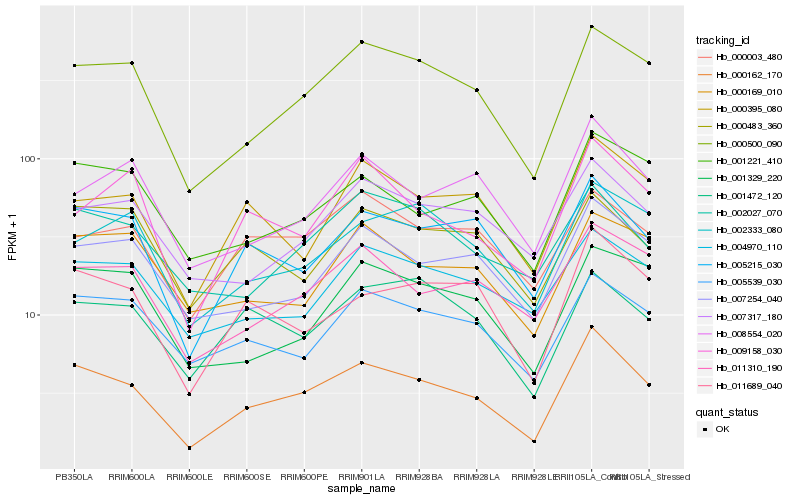
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 2 |
Hb_000500_090 |
0.093299729 |
- |
- |
nucleoside diphosphate kinase 2 [Hevea brasiliensis] |
| 3 |
Hb_005215_030 |
0.1068300966 |
- |
- |
FAD-dependent oxidoreductase family protein [Populus trichocarpa] |
| 4 |
Hb_011689_040 |
0.1100323868 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 5 |
Hb_007254_040 |
0.1171605116 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_011310_190 |
0.1182832842 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001221_410 |
0.1225840823 |
- |
- |
PREDICTED: gamma carbonic anhydrase-like 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005539_030 |
0.1231294918 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 9 |
Hb_000395_080 |
0.1267106579 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007317_180 |
0.1270012473 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 11 |
Hb_002027_070 |
0.1277010874 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 12 |
Hb_004970_110 |
0.1283835862 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 13 |
Hb_008554_020 |
0.1315624032 |
- |
- |
- |
| 14 |
Hb_001329_220 |
0.1320940755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000003_480 |
0.1340080274 |
- |
- |
PREDICTED: nuclear pore complex protein NUP35 [Jatropha curcas] |
| 16 |
Hb_002333_080 |
0.1352882742 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20-like [Populus euphratica] |
| 17 |
Hb_009158_030 |
0.1366341447 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 18 |
Hb_001472_120 |
0.1370265114 |
- |
- |
- |
| 19 |
Hb_000169_010 |
0.1373898197 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000483_360 |
0.1375977435 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |