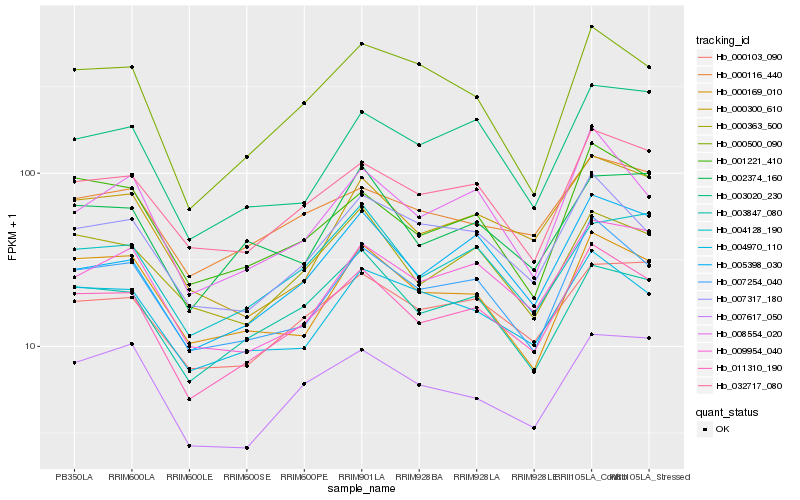
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007254_040 |
0.0631102962 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_032717_080 |
0.0671137599 |
- |
- |
gamma-soluble nsf attachment protein, putative [Ricinus communis] |
| 4 |
Hb_005398_030 |
0.0773858933 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase 1-like [Jatropha curcas] |
| 5 |
Hb_007317_180 |
0.0793358698 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 6 |
Hb_001221_410 |
0.0806621791 |
- |
- |
PREDICTED: gamma carbonic anhydrase-like 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000103_090 |
0.0809050627 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |
| 8 |
Hb_000116_440 |
0.0819919538 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 6-2 [Jatropha curcas] |
| 9 |
Hb_000363_500 |
0.0826811928 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004970_110 |
0.0835517601 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 11 |
Hb_003847_080 |
0.0866051969 |
- |
- |
PREDICTED: uncharacterized protein LOC105647792 isoform X3 [Jatropha curcas] |
| 12 |
Hb_008554_020 |
0.0869572885 |
- |
- |
- |
| 13 |
Hb_009954_040 |
0.0870045184 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 14 |
Hb_000300_610 |
0.0894991885 |
- |
- |
cytosolic copper/zinc superoxide dismutase [Hevea brasiliensis] |
| 15 |
Hb_004128_190 |
0.0919057447 |
- |
- |
PREDICTED: dirigent protein 17-like [Jatropha curcas] |
| 16 |
Hb_007617_050 |
0.0923680958 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 17 |
Hb_000169_010 |
0.0935361099 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_000500_090 |
0.095095549 |
- |
- |
nucleoside diphosphate kinase 2 [Hevea brasiliensis] |
| 19 |
Hb_003020_230 |
0.0956568541 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 20 |
Hb_002374_160 |
0.0959461782 |
- |
- |
PREDICTED: N-glycosylase/DNA lyase OGG1 isoform X1 [Jatropha curcas] |