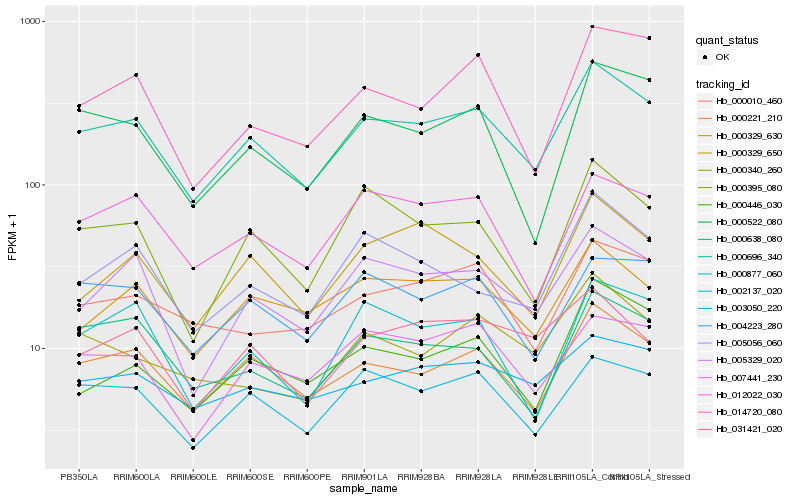
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 2 |
Hb_000221_210 |
0.0604922947 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_002137_020 |
0.0822842071 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_005329_020 |
0.0856098155 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 5 |
Hb_000329_630 |
0.0926092172 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 6 |
Hb_004223_280 |
0.0950118539 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 7 |
Hb_012022_030 |
0.0954644465 |
transcription factor |
TF Family: Alfin-like |
phd/F-box containing protein, putative [Ricinus communis] |
| 8 |
Hb_000395_080 |
0.0963895406 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007441_230 |
0.0975502977 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase [Jatropha curcas] |
| 10 |
Hb_003050_220 |
0.0975573511 |
- |
- |
hypothetical protein JCGZ_04601 [Jatropha curcas] |
| 11 |
Hb_000010_460 |
0.097959305 |
- |
- |
hypothetical protein EUGRSUZ_B001502, partial [Eucalyptus grandis] |
| 12 |
Hb_031421_020 |
0.099900696 |
- |
- |
ADP-ribose pyrophosphatase, putative [Ricinus communis] |
| 13 |
Hb_000638_080 |
0.1004417271 |
- |
- |
PREDICTED: uncharacterized protein LOC105632765 [Jatropha curcas] |
| 14 |
Hb_000522_080 |
0.1009885304 |
- |
- |
PREDICTED: uncharacterized protein LOC105632108 [Jatropha curcas] |
| 15 |
Hb_000329_650 |
0.1019289876 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 16 |
Hb_005056_060 |
0.1050735753 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000877_060 |
0.1056254425 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 18 |
Hb_000340_260 |
0.105966131 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 19 |
Hb_014720_080 |
0.1068752475 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 20 |
Hb_000446_030 |
0.1076517009 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |