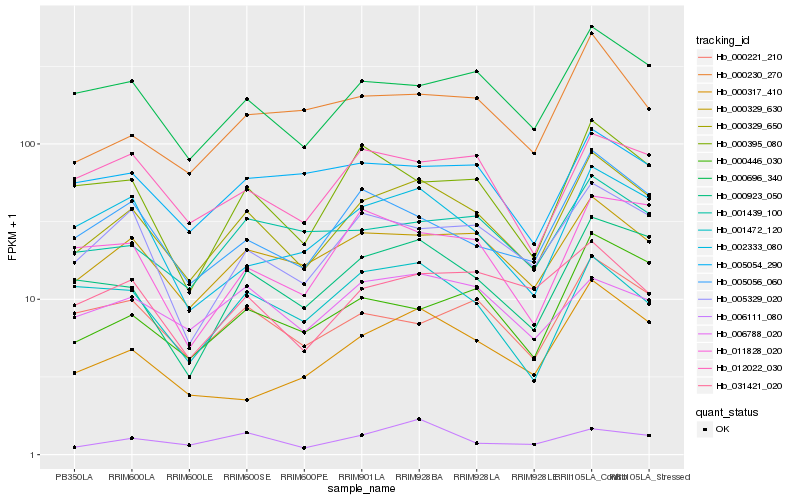
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_650 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 2 |
Hb_000329_630 |
0.0905709969 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 3 |
Hb_000923_050 |
0.0975014453 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000696_340 |
0.1019289876 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 5 |
Hb_005329_020 |
0.1089135066 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 6 |
Hb_005056_060 |
0.115250168 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 7 |
Hb_002333_080 |
0.1193369013 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20-like [Populus euphratica] |
| 8 |
Hb_000395_080 |
0.1207611751 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012022_030 |
0.1213698961 |
transcription factor |
TF Family: Alfin-like |
phd/F-box containing protein, putative [Ricinus communis] |
| 10 |
Hb_001439_100 |
0.1232636285 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 11 |
Hb_000446_030 |
0.1267874717 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_031421_020 |
0.1269864461 |
- |
- |
ADP-ribose pyrophosphatase, putative [Ricinus communis] |
| 13 |
Hb_000230_270 |
0.1273701237 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 14 |
Hb_000221_210 |
0.1273758676 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_000317_410 |
0.1280552826 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 16 |
Hb_006788_020 |
0.130264774 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 17 |
Hb_005054_290 |
0.1313503033 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit F [Jatropha curcas] |
| 18 |
Hb_006111_080 |
0.1319817232 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 19 |
Hb_001472_120 |
0.1329826217 |
- |
- |
- |
| 20 |
Hb_011828_020 |
0.13372822 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-10-like isoform X2 [Jatropha curcas] |