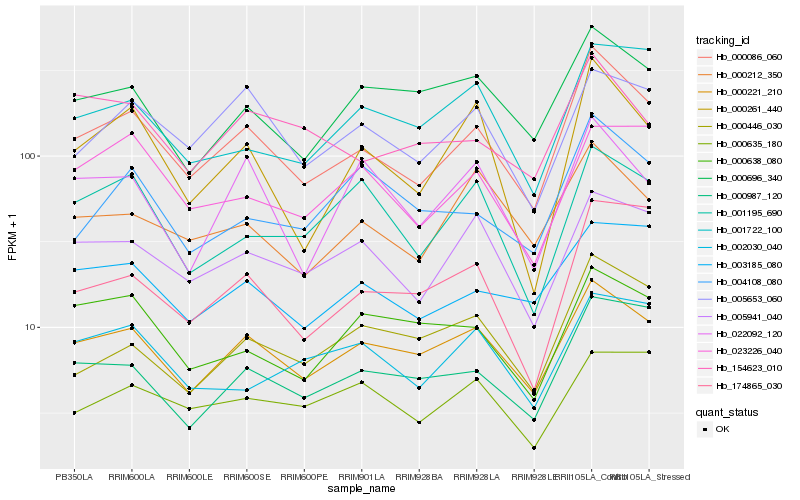
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_060 |
0.0 |
- |
- |
PREDICTED: TOM1-like protein 2 isoform X2 [Populus euphratica] |
| 2 |
Hb_000221_210 |
0.0874640438 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_005941_040 |
0.1164662411 |
- |
- |
PREDICTED: CASP-like protein 5A2 [Jatropha curcas] |
| 4 |
Hb_000261_440 |
0.1169809044 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000987_120 |
0.1190712968 |
- |
- |
PREDICTED: protein SIEL [Jatropha curcas] |
| 6 |
Hb_005653_060 |
0.121057237 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 7 |
Hb_000212_350 |
0.121651385 |
- |
- |
glucosamine-fructose-6-phosphate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_003185_080 |
0.1228225086 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_174865_030 |
0.1242906027 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 10 |
Hb_000696_340 |
0.1247129849 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 11 |
Hb_000446_030 |
0.1297672133 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_004108_080 |
0.131550366 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase [Vitis vinifera] |
| 13 |
Hb_001722_100 |
0.1339507776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_023226_040 |
0.1341609543 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_154623_010 |
0.1359856734 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001195_690 |
0.1360083623 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185 [Jatropha curcas] |
| 17 |
Hb_022092_120 |
0.1375726305 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 18 |
Hb_002030_040 |
0.1379798004 |
- |
- |
hypothetical protein JCGZ_18677 [Jatropha curcas] |
| 19 |
Hb_000638_080 |
0.1392208162 |
- |
- |
PREDICTED: uncharacterized protein LOC105632765 [Jatropha curcas] |
| 20 |
Hb_000635_180 |
0.139419875 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |