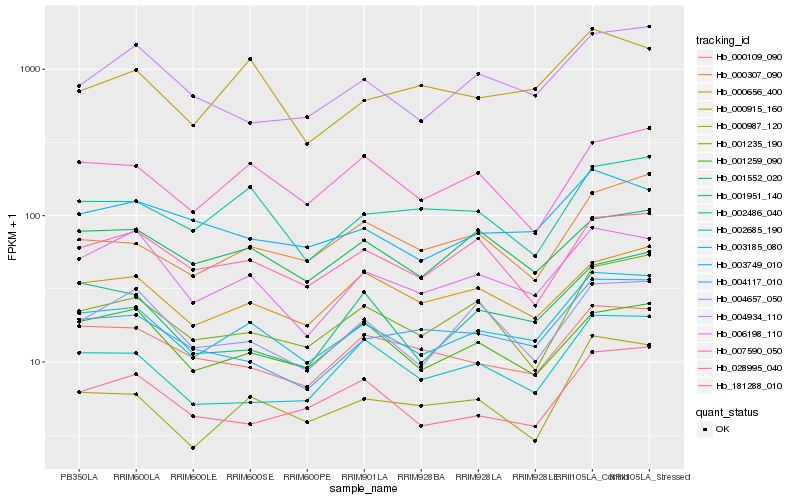
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003185_080 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_006198_110 |
0.0878869355 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000656_400 |
0.0900583271 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 4 |
Hb_003749_010 |
0.0914886825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004117_010 |
0.0938062905 |
- |
- |
60S ribosomal protein L10, mitochondrial, putative [Ricinus communis] |
| 6 |
Hb_001235_190 |
0.0944521994 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 7 |
Hb_028995_040 |
0.0946824552 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001552_020 |
0.0956785741 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 9 |
Hb_002486_040 |
0.0958691583 |
- |
- |
- |
| 10 |
Hb_000109_090 |
0.0963851726 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 11 |
Hb_007590_050 |
0.0972999322 |
- |
- |
NudC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_181288_010 |
0.1011663165 |
- |
- |
rac-GTP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000987_120 |
0.1014772294 |
- |
- |
PREDICTED: protein SIEL [Jatropha curcas] |
| 14 |
Hb_000915_160 |
0.1015030523 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 15 |
Hb_004934_110 |
0.1017214217 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 16 |
Hb_001259_090 |
0.1018226625 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 17 |
Hb_000307_090 |
0.1018341622 |
- |
- |
hypothetical protein CICLE_v10002475mg [Citrus clementina] |
| 18 |
Hb_002685_190 |
0.1027142777 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004657_050 |
0.1033423743 |
- |
- |
PREDICTED: tRNA-dihydrouridine(16/17) synthase [NAD(P)(+)]-like [Jatropha curcas] |
| 20 |
Hb_001951_140 |
0.1058438211 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X1 [Jatropha curcas] |