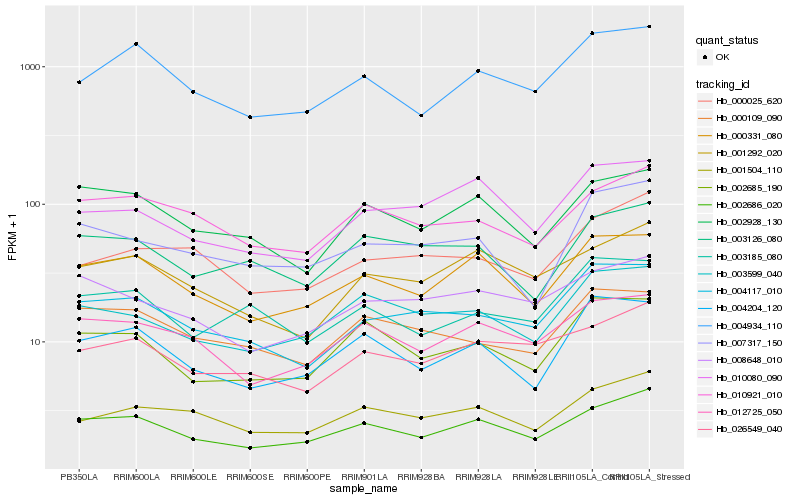
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_010 |
0.0 |
- |
- |
60S ribosomal protein L10, mitochondrial, putative [Ricinus communis] |
| 2 |
Hb_000109_090 |
0.0816885683 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 3 |
Hb_003185_080 |
0.0938062905 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_026549_040 |
0.0971964432 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008648_010 |
0.0981395133 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Jatropha curcas] |
| 6 |
Hb_003599_040 |
0.0988735937 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000331_080 |
0.0989851956 |
- |
- |
PREDICTED: UPF0235 protein At5g63440 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_002686_020 |
0.0993169729 |
- |
- |
galactose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_010080_090 |
0.0996811515 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 [Jatropha curcas] |
| 10 |
Hb_002685_190 |
0.1001029271 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002928_130 |
0.1002224804 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 12 |
Hb_000025_620 |
0.1010809492 |
- |
- |
hypothetical protein POPTR_0009s05200g [Populus trichocarpa] |
| 13 |
Hb_001292_020 |
0.1010903328 |
- |
- |
PREDICTED: keratin, type II cytoskeletal 5 isoform X1 [Sesamum indicum] |
| 14 |
Hb_004934_110 |
0.1019934311 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 15 |
Hb_012725_050 |
0.102602173 |
transcription factor |
TF Family: SET |
set domain protein, putative [Ricinus communis] |
| 16 |
Hb_003126_080 |
0.1026979879 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 17 |
Hb_004204_120 |
0.1028300939 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 18 |
Hb_010921_010 |
0.1031785691 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 19 |
Hb_007317_150 |
0.1044256443 |
- |
- |
PREDICTED: 40S ribosomal protein S25 [Jatropha curcas] |
| 20 |
Hb_001504_110 |
0.1050814496 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |