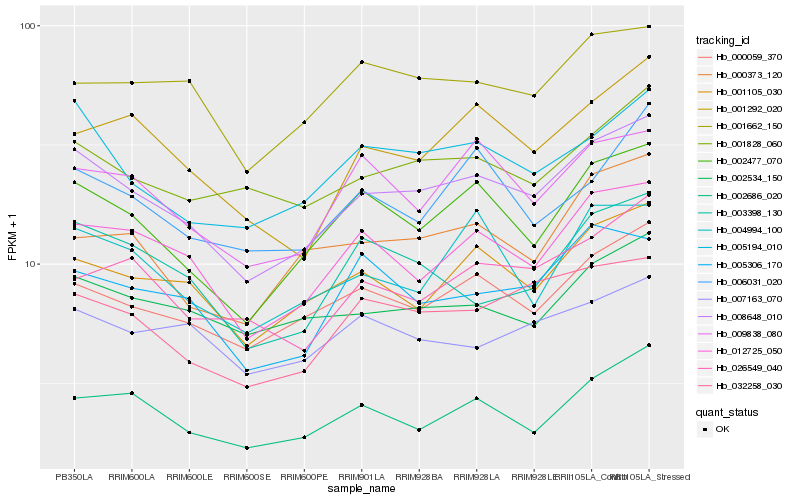
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008648_010 |
0.0 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Jatropha curcas] |
| 2 |
Hb_002477_070 |
0.0671489841 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 3 |
Hb_000059_370 |
0.0697101748 |
- |
- |
PREDICTED: U6 snRNA phosphodiesterase [Jatropha curcas] |
| 4 |
Hb_012725_050 |
0.0705556229 |
transcription factor |
TF Family: SET |
set domain protein, putative [Ricinus communis] |
| 5 |
Hb_005194_010 |
0.0721619016 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 6 |
Hb_001105_030 |
0.0748921425 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 7 |
Hb_032258_030 |
0.0758217751 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 8 |
Hb_002686_020 |
0.0817526221 |
- |
- |
galactose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001828_060 |
0.0833262719 |
desease resistance |
Gene Name: AAA_31 |
nucleotide-binding protein, putative [Ricinus communis] |
| 10 |
Hb_002534_150 |
0.0839575824 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_001292_020 |
0.0880805766 |
- |
- |
PREDICTED: keratin, type II cytoskeletal 5 isoform X1 [Sesamum indicum] |
| 12 |
Hb_009838_080 |
0.0889486385 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001662_150 |
0.0892533642 |
- |
- |
Scaffold attachment factor B1 [Medicago truncatula] |
| 14 |
Hb_003398_130 |
0.0910606543 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL30 [Gossypium raimondii] |
| 15 |
Hb_026549_040 |
0.0917846247 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000373_120 |
0.0925176456 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 17 |
Hb_005306_170 |
0.0927690911 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 18 |
Hb_004994_100 |
0.0928779015 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 19 |
Hb_006031_020 |
0.0955070297 |
- |
- |
PREDICTED: protein FLX-like 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007163_070 |
0.0956247596 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |