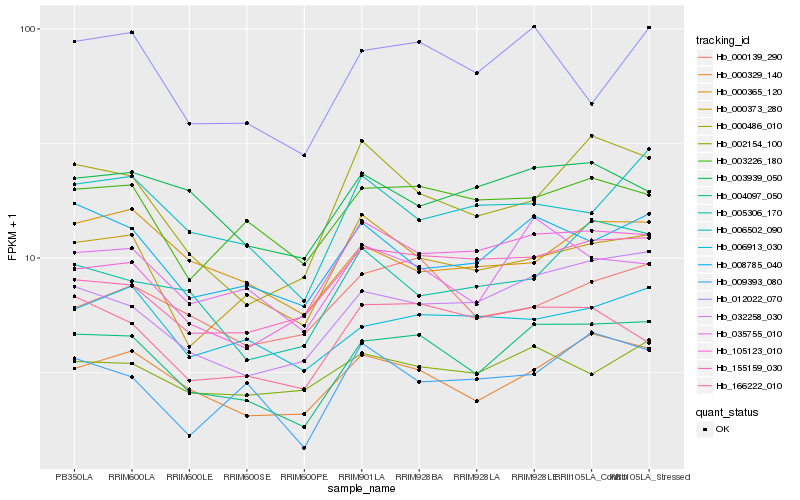
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004097_050 |
0.0 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 2 |
Hb_032258_030 |
0.082222674 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 3 |
Hb_000329_140 |
0.0874861418 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 4 |
Hb_008785_040 |
0.0996392665 |
- |
- |
PREDICTED: DNA repair protein UVH3 [Jatropha curcas] |
| 5 |
Hb_035755_010 |
0.1004681132 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_012022_070 |
0.103882772 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 7 |
Hb_000373_280 |
0.1052114511 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_105123_010 |
0.1060313889 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 9 |
Hb_003226_180 |
0.1063555742 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 10 |
Hb_155159_030 |
0.1085161807 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_166222_010 |
0.1085700853 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 12 |
Hb_000486_010 |
0.1087832503 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006913_030 |
0.1093503018 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 14 |
Hb_006502_090 |
0.1161083907 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 15 |
Hb_002154_100 |
0.1164243479 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 16 |
Hb_003939_050 |
0.1182499369 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_005306_170 |
0.1186595407 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 18 |
Hb_000139_290 |
0.1205982208 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000365_120 |
0.1226834945 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_009393_080 |
0.123991504 |
- |
- |
plant organelle RNA recognition domain protein [Medicago truncatula] |