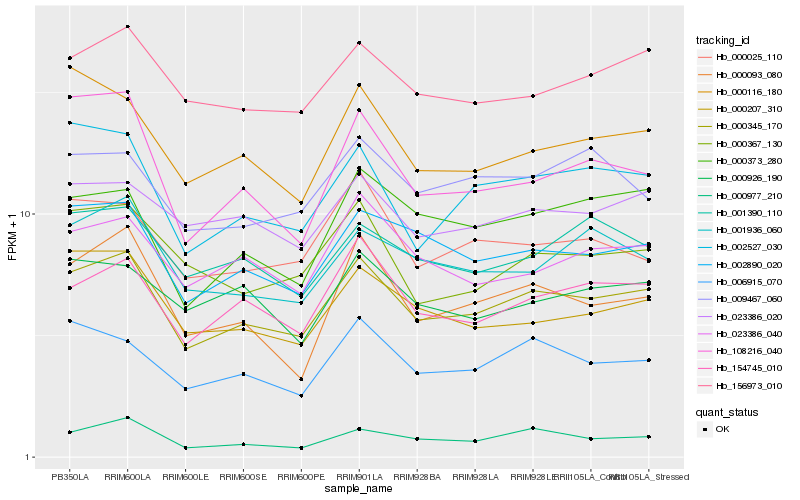
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_170 |
0.0 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002890_020 |
0.0638371942 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_154745_010 |
0.0659447493 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_156973_010 |
0.070616161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006915_070 |
0.0716437673 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 6 |
Hb_002527_030 |
0.0727721241 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1-like [Jatropha curcas] |
| 7 |
Hb_023386_040 |
0.0764704593 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 8 |
Hb_023386_020 |
0.0765484182 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000207_310 |
0.0774392981 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 10 |
Hb_000977_210 |
0.0778884063 |
- |
- |
chromatin assembly factor 1, subunit A, putative [Ricinus communis] |
| 11 |
Hb_001936_060 |
0.0782410744 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 12 |
Hb_000373_280 |
0.0783435811 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000025_110 |
0.0803794196 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 14 |
Hb_001390_110 |
0.0805178197 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 15 |
Hb_000116_180 |
0.0812050794 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 16 |
Hb_000367_130 |
0.0819687117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000093_080 |
0.0825250983 |
- |
- |
hypothetical protein POPTR_0008s09730g [Populus trichocarpa] |
| 18 |
Hb_009467_060 |
0.0849098471 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000926_190 |
0.0849594639 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_108216_040 |
0.085759623 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |