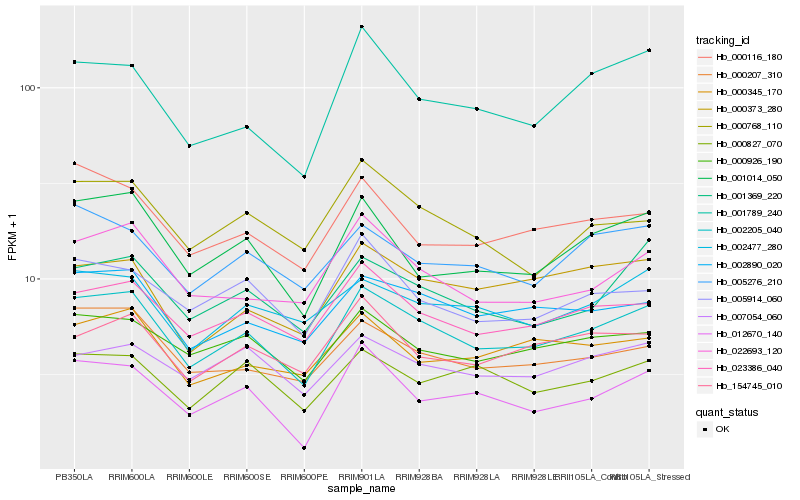
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_040 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_023386_040 |
0.0701363871 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 3 |
Hb_002890_020 |
0.0720376739 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001014_050 |
0.0760207926 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 5 |
Hb_000926_190 |
0.0762539529 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000207_310 |
0.0789227819 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 7 |
Hb_022693_120 |
0.0795149976 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 8 |
Hb_001369_220 |
0.0807084104 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001789_240 |
0.0812981368 |
- |
- |
hypothetical protein CICLE_v10032221mg [Citrus clementina] |
| 10 |
Hb_000116_180 |
0.0864683578 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 11 |
Hb_000373_280 |
0.0868383754 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005276_210 |
0.0868510281 |
- |
- |
PREDICTED: DNA repair helicase UVH6 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002477_280 |
0.0871101566 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-2 [Jatropha curcas] |
| 14 |
Hb_005914_060 |
0.0873347919 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 15 |
Hb_012670_140 |
0.0876551406 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g28690, mitochondrial [Jatropha curcas] |
| 16 |
Hb_154745_010 |
0.0884287762 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 17 |
Hb_007054_060 |
0.0889485205 |
- |
- |
- |
| 18 |
Hb_000827_070 |
0.0891900466 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 19 |
Hb_000345_170 |
0.0892475632 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000768_110 |
0.0894386862 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |