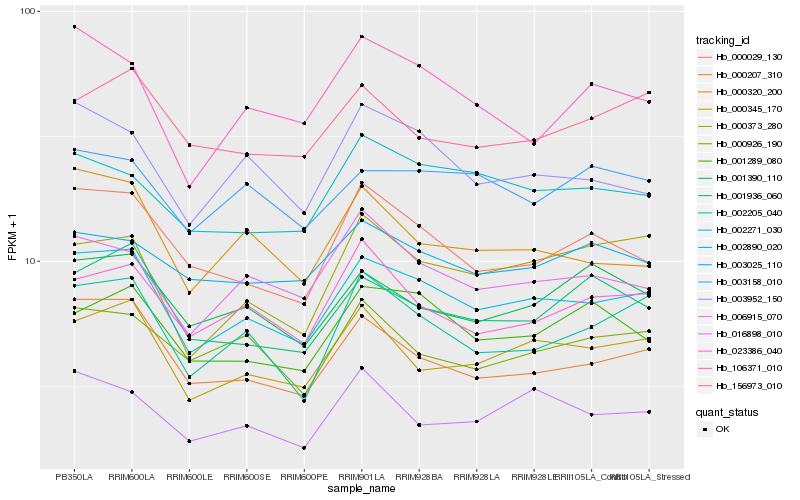
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_020 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000207_310 |
0.061115684 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 3 |
Hb_000345_170 |
0.0638371942 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002205_040 |
0.0720376739 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000029_130 |
0.07261892 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_016898_010 |
0.0740247008 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 7 |
Hb_003952_150 |
0.074400192 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 8 |
Hb_023386_040 |
0.0775838843 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 9 |
Hb_001936_060 |
0.0777121701 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 10 |
Hb_002271_030 |
0.0783714521 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 11 |
Hb_000320_200 |
0.0788052549 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 12 |
Hb_001289_080 |
0.0803158654 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 13 |
Hb_001390_110 |
0.0809578492 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 14 |
Hb_106371_010 |
0.0820880844 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_000373_280 |
0.0822050894 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000926_190 |
0.0822642362 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_003158_010 |
0.0828310056 |
- |
- |
hypothetical protein JCGZ_17361 [Jatropha curcas] |
| 18 |
Hb_003025_110 |
0.0833454131 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006915_070 |
0.0834738681 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 20 |
Hb_156973_010 |
0.0836874964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |