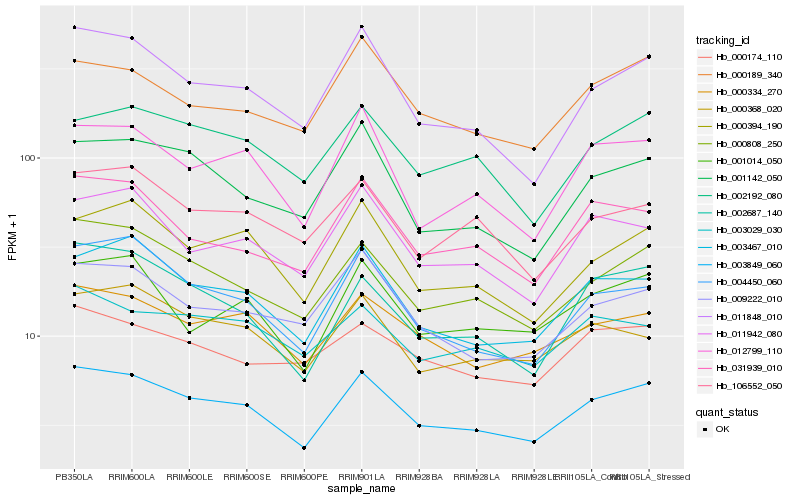
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003849_060 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000808_250 |
0.0555062961 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 3 |
Hb_000334_270 |
0.0635983557 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003467_010 |
0.0668436193 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 5 |
Hb_011848_010 |
0.0732253759 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 6 |
Hb_000394_190 |
0.0744571979 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002192_080 |
0.0744824045 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 8 |
Hb_031939_010 |
0.0750105894 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001014_050 |
0.075104887 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 10 |
Hb_004450_060 |
0.0764446042 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002687_140 |
0.0772840464 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 12 |
Hb_000189_340 |
0.0774508058 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 13 |
Hb_012799_110 |
0.0774818277 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 14 |
Hb_009222_010 |
0.0781068519 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_000368_020 |
0.0781203427 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_011942_080 |
0.0781574989 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 17 |
Hb_000174_110 |
0.0806957967 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 18 |
Hb_106552_050 |
0.0808370544 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 19 |
Hb_003029_030 |
0.0808895606 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001142_050 |
0.081255435 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |