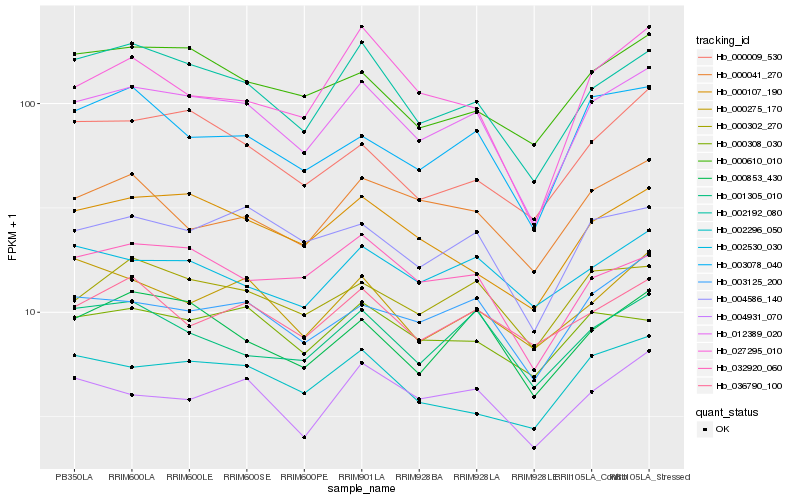
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012389_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 2 |
Hb_002192_080 |
0.0579304775 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 3 |
Hb_004586_140 |
0.0705351561 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 4 |
Hb_003125_200 |
0.0709888512 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 5 |
Hb_004931_070 |
0.0761181285 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000302_270 |
0.0763050597 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000853_430 |
0.0769559672 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 8 |
Hb_000107_190 |
0.0772145974 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 9 |
Hb_032920_060 |
0.077591488 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.1 [Jatropha curcas] |
| 10 |
Hb_002296_050 |
0.0828813694 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 11 |
Hb_001305_010 |
0.0830203683 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003078_040 |
0.0845674268 |
- |
- |
PREDICTED: hit family protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000610_010 |
0.085916204 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_000041_270 |
0.0865642531 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 15 |
Hb_000308_030 |
0.0866687305 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 16 |
Hb_002530_030 |
0.088860838 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 17 |
Hb_000009_530 |
0.0893436808 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 18 |
Hb_000275_170 |
0.0894358115 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 19 |
Hb_027295_010 |
0.0896473199 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 20 |
Hb_036790_100 |
0.0896982799 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |