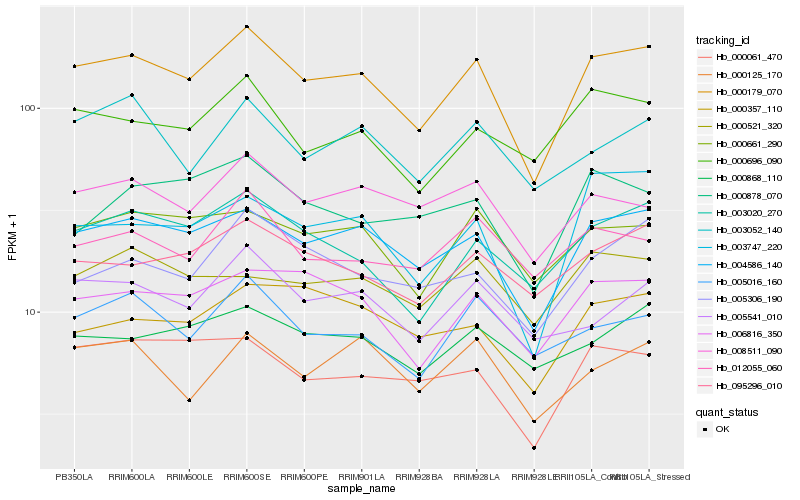
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_070 |
0.0 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 2 |
Hb_004586_140 |
0.0505042565 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 3 |
Hb_006816_350 |
0.0811999425 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 4 |
Hb_003020_270 |
0.0818715882 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 5 |
Hb_000661_290 |
0.0856674828 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_095296_010 |
0.0881633194 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 7 |
Hb_000696_090 |
0.0905964135 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 8 |
Hb_000061_470 |
0.0910575305 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 9 |
Hb_008511_090 |
0.0918656678 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 10 |
Hb_005016_160 |
0.0928380354 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 11 |
Hb_003747_220 |
0.0938569642 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000868_110 |
0.0939128609 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 13 |
Hb_005306_190 |
0.0939147197 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 14 |
Hb_005541_010 |
0.0942141256 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 15 |
Hb_000125_170 |
0.0964653793 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 16 |
Hb_003052_140 |
0.0966653561 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 17 |
Hb_000357_110 |
0.097780792 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012055_060 |
0.0979880951 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000878_070 |
0.1009082318 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 20 |
Hb_000521_320 |
0.1014543928 |
- |
- |
hypothetical protein POPTR_0001s23520g [Populus trichocarpa] |