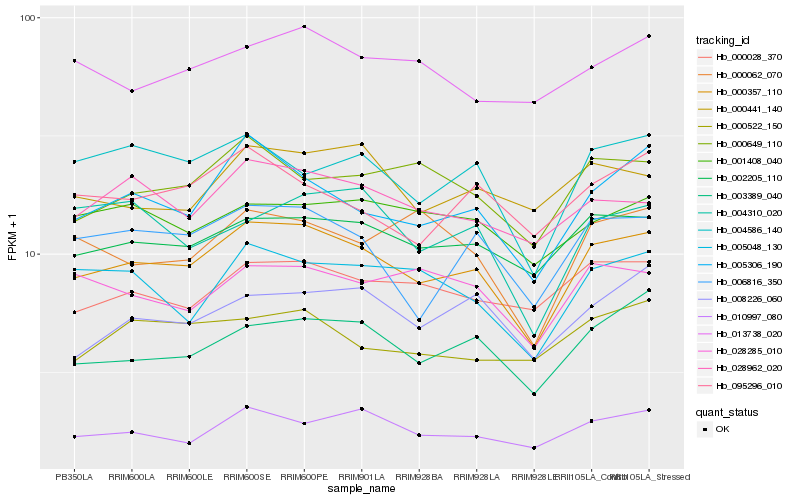
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000357_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002205_110 |
0.071021691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005306_190 |
0.0736395601 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 4 |
Hb_003389_040 |
0.0765025807 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 5 |
Hb_006816_350 |
0.0784946677 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 6 |
Hb_010997_080 |
0.0800764825 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_004586_140 |
0.0816905593 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 8 |
Hb_028962_020 |
0.0834882317 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 9 |
Hb_005048_130 |
0.0838621139 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 10 |
Hb_000649_110 |
0.0866588489 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 11 |
Hb_000028_370 |
0.0869892142 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 12 |
Hb_008226_060 |
0.0891827944 |
- |
- |
PREDICTED: folic acid synthesis protein fol1-like [Jatropha curcas] |
| 13 |
Hb_028285_010 |
0.0905302676 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 14 |
Hb_001408_040 |
0.0921223671 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 15 |
Hb_095296_010 |
0.0924659027 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 16 |
Hb_000441_140 |
0.092880874 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-7-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_013738_020 |
0.0940207844 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 18 |
Hb_004310_020 |
0.0940348356 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000522_150 |
0.0948263075 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 20 |
Hb_000062_070 |
0.0948274375 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |