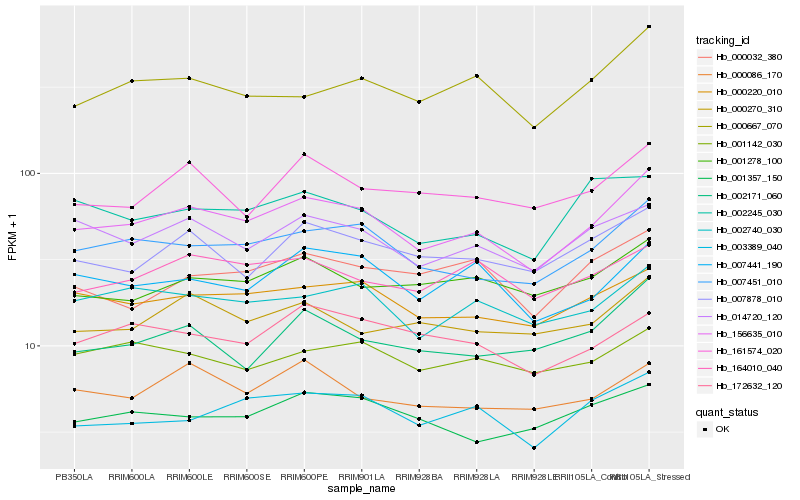
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156635_010 |
0.0 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 2 |
Hb_007451_010 |
0.0567205769 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 3 |
Hb_000220_010 |
0.0722004252 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 4 |
Hb_014720_120 |
0.0753323647 |
- |
- |
Phosphoenolpyruvate/phosphate translocator 1, chloroplastic -like protein [Gossypium arboreum] |
| 5 |
Hb_002740_030 |
0.0765080291 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003389_040 |
0.0779955689 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 7 |
Hb_007441_190 |
0.0818026523 |
- |
- |
hypothetical protein CISIN_1g023046mg [Citrus sinensis] |
| 8 |
Hb_007878_010 |
0.0828629349 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 9 |
Hb_000667_070 |
0.0848203925 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 10 |
Hb_000270_310 |
0.0857171687 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 11 |
Hb_001278_100 |
0.0867526873 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 12 |
Hb_161574_020 |
0.0872227519 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_000086_170 |
0.0872643353 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 14 |
Hb_164010_040 |
0.0887467495 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001357_150 |
0.0902119297 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 16 |
Hb_002171_060 |
0.0902145576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_172632_120 |
0.0918949619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001142_030 |
0.0923141868 |
- |
- |
PREDICTED: uncharacterized protein LOC105640023 [Jatropha curcas] |
| 19 |
Hb_000032_380 |
0.093803694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002245_030 |
0.0942713898 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |