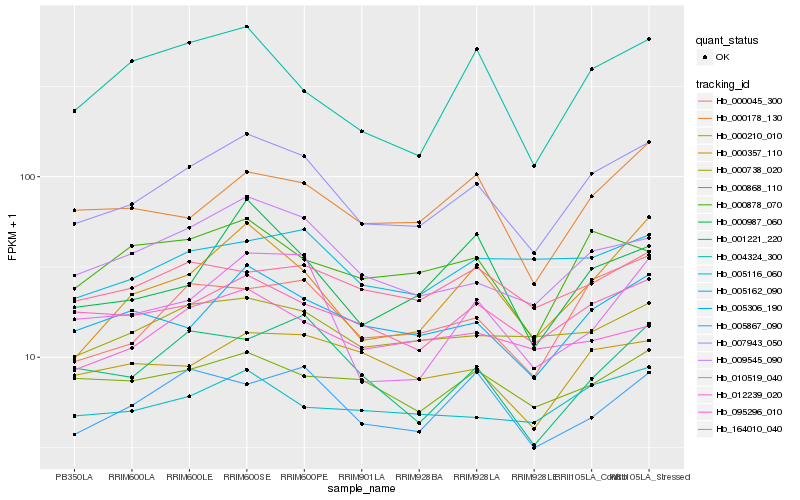
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_050 |
0.0 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 2 |
Hb_000045_300 |
0.0882005483 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 3 |
Hb_009545_090 |
0.0885635877 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 4 |
Hb_005306_190 |
0.0887213592 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 5 |
Hb_000210_010 |
0.0935268043 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 6 |
Hb_095296_010 |
0.0967507206 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 7 |
Hb_010519_040 |
0.1093809977 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 8 |
Hb_000738_020 |
0.1112355998 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000178_130 |
0.1112382729 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 10 |
Hb_005116_060 |
0.1123173282 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005867_090 |
0.1132925309 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 12 |
Hb_000357_110 |
0.113374572 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000987_060 |
0.1159372475 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 14 |
Hb_001221_220 |
0.1167681898 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 15 |
Hb_012239_020 |
0.1171434264 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 16 |
Hb_000878_070 |
0.1174776804 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 17 |
Hb_004324_300 |
0.1181901673 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 18 |
Hb_000868_110 |
0.1193724989 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 19 |
Hb_005162_090 |
0.1194475863 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 20 |
Hb_164010_040 |
0.1215994393 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |