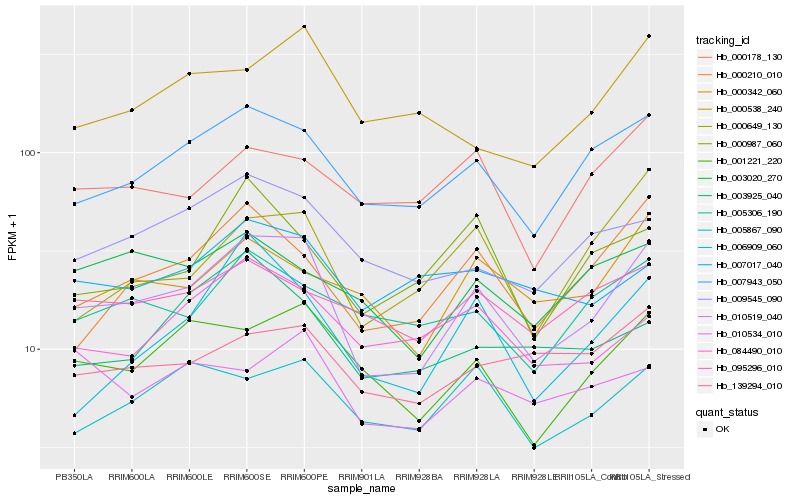
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010519_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 2 |
Hb_007943_050 |
0.1093809977 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_001221_220 |
0.1224434254 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 4 |
Hb_000210_010 |
0.1342759843 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 5 |
Hb_009545_090 |
0.1365704117 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 6 |
Hb_005867_090 |
0.1415578444 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 7 |
Hb_000342_060 |
0.1454556817 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 8 |
Hb_000649_130 |
0.1457590869 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 9 |
Hb_005306_190 |
0.1463591214 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 10 |
Hb_139294_010 |
0.1468793272 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_006909_060 |
0.1476856311 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 12 |
Hb_000538_240 |
0.1480792016 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 13 |
Hb_000178_130 |
0.149510822 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 14 |
Hb_084490_010 |
0.150569273 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003020_270 |
0.1512211064 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 16 |
Hb_095296_010 |
0.1515377926 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 17 |
Hb_000987_060 |
0.1526495407 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 18 |
Hb_007017_040 |
0.1545736408 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 19 |
Hb_010534_010 |
0.1572155199 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 20 |
Hb_003925_040 |
0.1577868495 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |