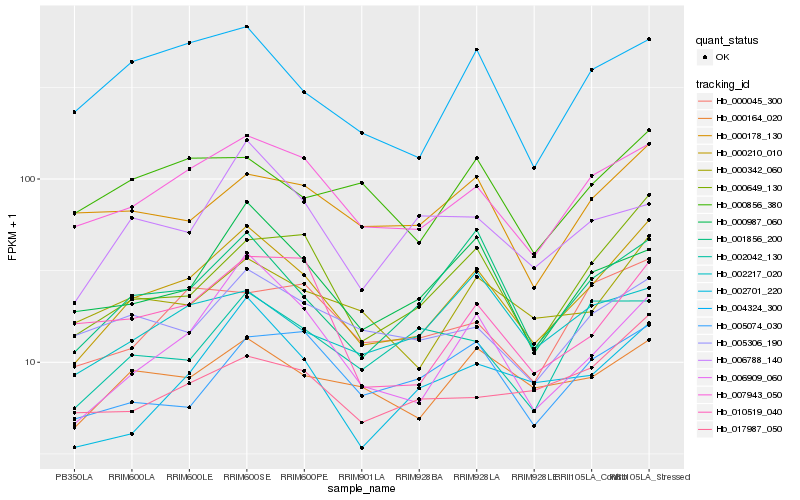
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_010 |
0.0 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 2 |
Hb_007943_050 |
0.0935268043 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_000649_130 |
0.1063107884 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 4 |
Hb_006909_060 |
0.1102410823 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 5 |
Hb_001856_200 |
0.1111336491 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 6 |
Hb_000342_060 |
0.1182254971 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 7 |
Hb_000164_020 |
0.123824692 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 8 |
Hb_004324_300 |
0.1248039908 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 9 |
Hb_002701_220 |
0.1253447632 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 10 |
Hb_000987_060 |
0.1259882304 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 11 |
Hb_010519_040 |
0.1342759843 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 12 |
Hb_017987_050 |
0.135105518 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_005306_190 |
0.1353833312 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 14 |
Hb_000178_130 |
0.1354338253 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 15 |
Hb_000045_300 |
0.136882798 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 16 |
Hb_000856_380 |
0.1414106808 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 17 |
Hb_005074_030 |
0.1429048482 |
- |
- |
- |
| 18 |
Hb_002217_020 |
0.1431762676 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006788_140 |
0.1471822704 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 20 |
Hb_002042_130 |
0.1486869026 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |