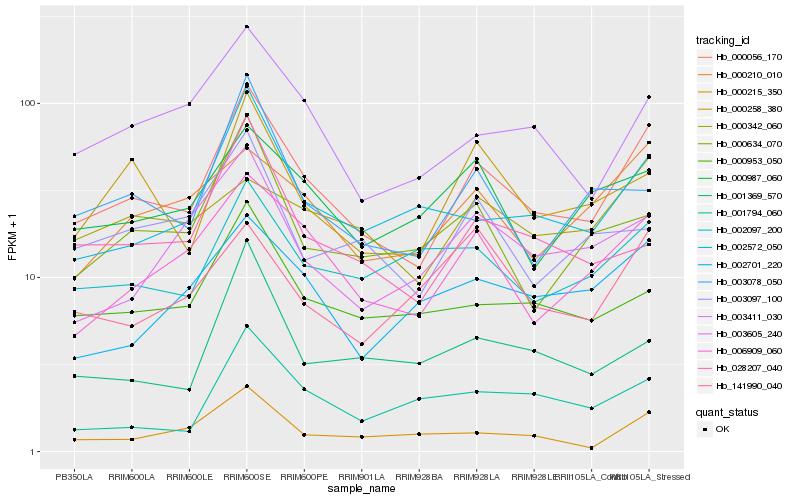
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006909_060 |
0.1243939183 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 3 |
Hb_003411_030 |
0.1416059088 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |
| 4 |
Hb_002572_050 |
0.1422522593 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_000258_380 |
0.1468003536 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |
| 6 |
Hb_000953_050 |
0.1531998771 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 7 |
Hb_000210_010 |
0.1534233019 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 8 |
Hb_002097_200 |
0.1542791852 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_003078_050 |
0.1575077748 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 10 |
Hb_002701_220 |
0.157638696 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 11 |
Hb_000987_060 |
0.15965773 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 12 |
Hb_028207_040 |
0.161096617 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 13 |
Hb_000215_350 |
0.1612262551 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_003605_240 |
0.1614288448 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 15 |
Hb_000634_070 |
0.1628401152 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 16 |
Hb_003097_100 |
0.1709308459 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 17 |
Hb_001794_060 |
0.172735162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001369_570 |
0.175309514 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 19 |
Hb_141990_040 |
0.175366561 |
- |
- |
Protein SCAR2, putative [Ricinus communis] |
| 20 |
Hb_000342_060 |
0.1754049405 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |