| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
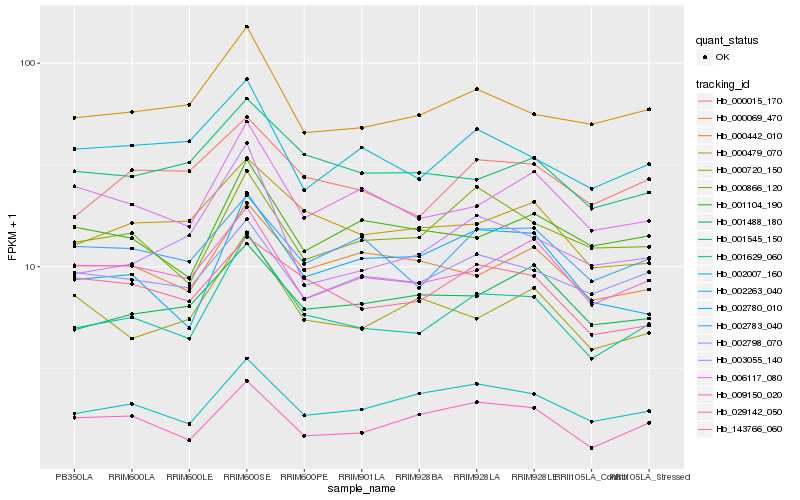
Hb_001629_060 |
0.0 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002007_160 |
0.0837835571 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 3 |
Hb_006117_080 |
0.08807738 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000442_010 |
0.0881063416 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 5 |
Hb_000479_070 |
0.0905662528 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 6 |
Hb_009150_020 |
0.0918453434 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 7 |
Hb_000015_170 |
0.0948603444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_143766_060 |
0.0962750797 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 9 |
Hb_001545_150 |
0.0967573429 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 10 |
Hb_002783_040 |
0.0978414651 |
transcription factor |
TF Family: IWS1 |
- |
| 11 |
Hb_002798_070 |
0.0991826423 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 12 |
Hb_000069_470 |
0.099842679 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003055_140 |
0.1007796279 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 14 |
Hb_001488_180 |
0.1009257104 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000866_120 |
0.1011245134 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 16 |
Hb_029142_050 |
0.101290882 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_002780_010 |
0.1017737937 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 18 |
Hb_001104_190 |
0.1024501206 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 19 |
Hb_000720_150 |
0.1027348513 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002263_040 |
0.1028948246 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |