| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
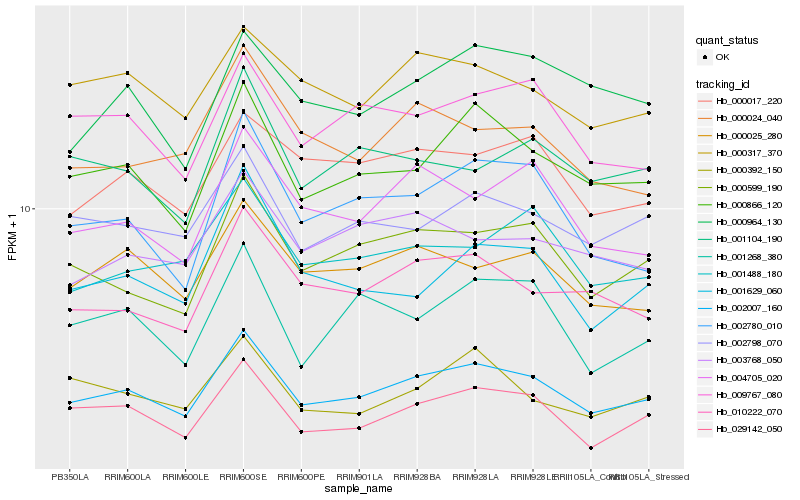
Hb_002007_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 2 |
Hb_000866_120 |
0.0561875748 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 3 |
Hb_000599_190 |
0.0662133205 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 4 |
Hb_029142_050 |
0.0735276353 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_002780_010 |
0.0737927708 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 6 |
Hb_010222_070 |
0.0747371056 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002798_070 |
0.077734422 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 8 |
Hb_009767_080 |
0.0786421982 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 9 |
Hb_000025_280 |
0.0787550506 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 10 |
Hb_004705_020 |
0.0796175958 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 11 |
Hb_001268_380 |
0.0828537118 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 12 |
Hb_001629_060 |
0.0837835571 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000017_220 |
0.0849263391 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 14 |
Hb_000317_370 |
0.0860615745 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 15 |
Hb_000024_040 |
0.0863105637 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 16 |
Hb_001488_180 |
0.0863841143 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001104_190 |
0.0879153781 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 18 |
Hb_000964_130 |
0.0882486518 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 19 |
Hb_003768_050 |
0.0901184426 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000392_150 |
0.0901358936 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g74580 [Jatropha curcas] |