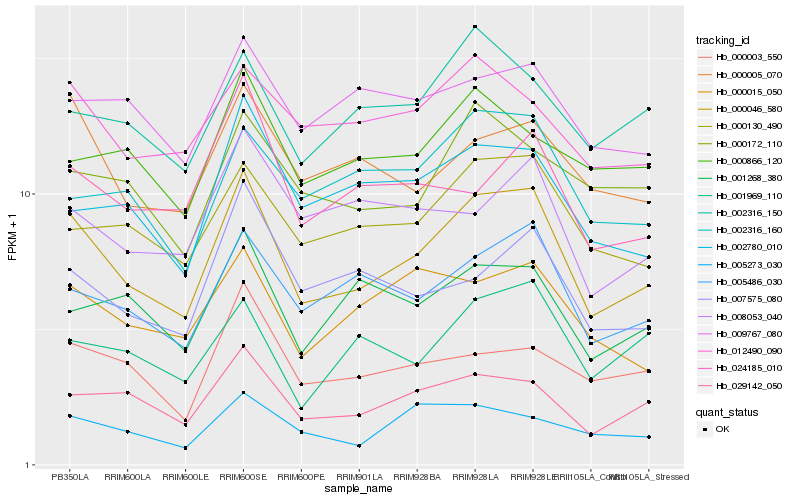
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_580 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 2 |
Hb_029142_050 |
0.0986365792 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_005486_030 |
0.1011552791 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 4 |
Hb_000130_490 |
0.1074860924 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000005_070 |
0.1147440901 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000015_050 |
0.1163522878 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 7 |
Hb_002780_010 |
0.1189755719 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 8 |
Hb_002316_150 |
0.1209773856 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 9 |
Hb_002316_160 |
0.1211157508 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 10 |
Hb_012490_090 |
0.1211387403 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 11 |
Hb_001268_380 |
0.1212148117 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 12 |
Hb_001969_110 |
0.1213083476 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000172_110 |
0.1224924266 |
- |
- |
- |
| 14 |
Hb_005273_030 |
0.1225097281 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 15 |
Hb_007575_080 |
0.1231058511 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 16 |
Hb_000003_550 |
0.1231370046 |
- |
- |
- |
| 17 |
Hb_024185_010 |
0.1245718219 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009767_080 |
0.1253762463 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 19 |
Hb_000866_120 |
0.1269040319 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 20 |
Hb_008053_040 |
0.1272083518 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |