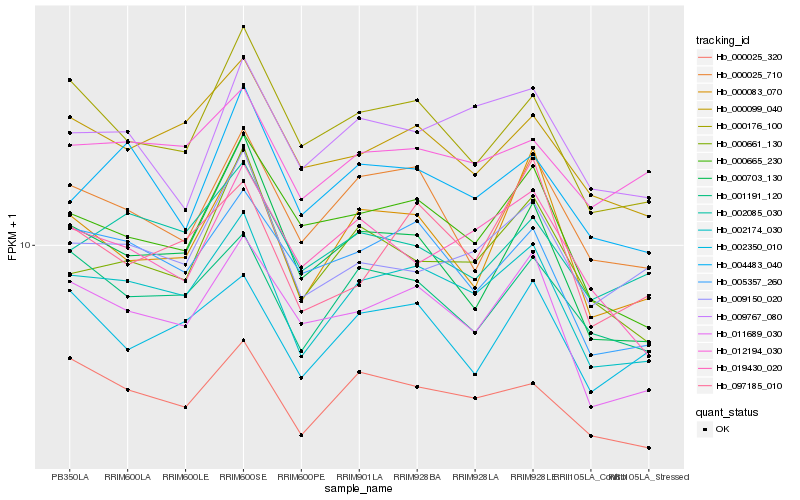
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002174_030 |
0.0 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 2 |
Hb_011689_030 |
0.0631401576 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 3 |
Hb_000025_320 |
0.0641001699 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 4 |
Hb_005357_260 |
0.0666760135 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 5 |
Hb_001191_120 |
0.0684418713 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000099_040 |
0.0706470542 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000176_100 |
0.0734362245 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 8 |
Hb_000665_230 |
0.0734701741 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_000025_710 |
0.0767592367 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 10 |
Hb_000083_070 |
0.0777185974 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000661_130 |
0.0786355935 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 12 |
Hb_012194_030 |
0.0789089067 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004483_040 |
0.0794104606 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 14 |
Hb_000703_130 |
0.081027742 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 15 |
Hb_009150_020 |
0.0813940944 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 16 |
Hb_002085_030 |
0.0817024786 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002350_010 |
0.0817124391 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 18 |
Hb_097185_010 |
0.0820082894 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 19 |
Hb_009767_080 |
0.0828753676 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 20 |
Hb_019430_020 |
0.0833121558 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |