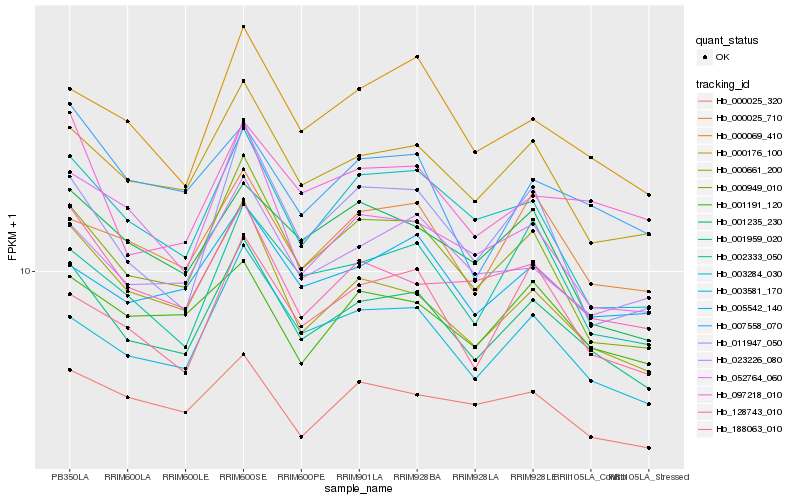
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000176_100 |
0.0523902819 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 3 |
Hb_000949_010 |
0.0607986975 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_003581_170 |
0.0617592752 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000661_200 |
0.0648035903 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000069_410 |
0.0709532845 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 7 |
Hb_128743_010 |
0.0712222541 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 8 |
Hb_023226_080 |
0.0718417083 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003284_030 |
0.0736052286 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000025_710 |
0.074661754 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 11 |
Hb_188063_010 |
0.0777366729 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 12 |
Hb_005542_140 |
0.0797696125 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 13 |
Hb_002333_050 |
0.0797962107 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001191_120 |
0.0803486481 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 15 |
Hb_097218_010 |
0.0810770117 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001235_230 |
0.0812318977 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 17 |
Hb_007558_070 |
0.0823256218 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011947_050 |
0.0823459964 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 19 |
Hb_000025_320 |
0.0832366372 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 20 |
Hb_052764_060 |
0.0837110578 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |