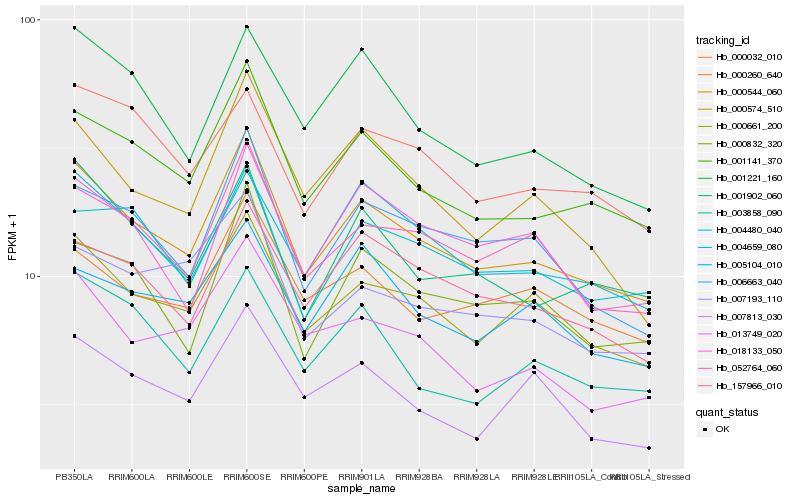
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000544_060 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 2 |
Hb_001141_370 |
0.046695767 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 3 |
Hb_000661_200 |
0.0559104541 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 4 |
Hb_052764_060 |
0.0644689893 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_006663_040 |
0.0711874805 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001902_060 |
0.0741762558 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000832_320 |
0.0751173948 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_004659_080 |
0.0770428168 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 9 |
Hb_157966_010 |
0.077253229 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 10 |
Hb_000574_510 |
0.0790504889 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_013749_020 |
0.0801315627 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000260_640 |
0.0803643625 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 13 |
Hb_001221_160 |
0.0821082877 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 14 |
Hb_007813_030 |
0.0823663474 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 15 |
Hb_004480_040 |
0.083569983 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005104_010 |
0.085384918 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 17 |
Hb_000032_010 |
0.0854019691 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 18 |
Hb_003858_090 |
0.0855213715 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_018133_050 |
0.0855633634 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 20 |
Hb_007193_110 |
0.0860286882 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |