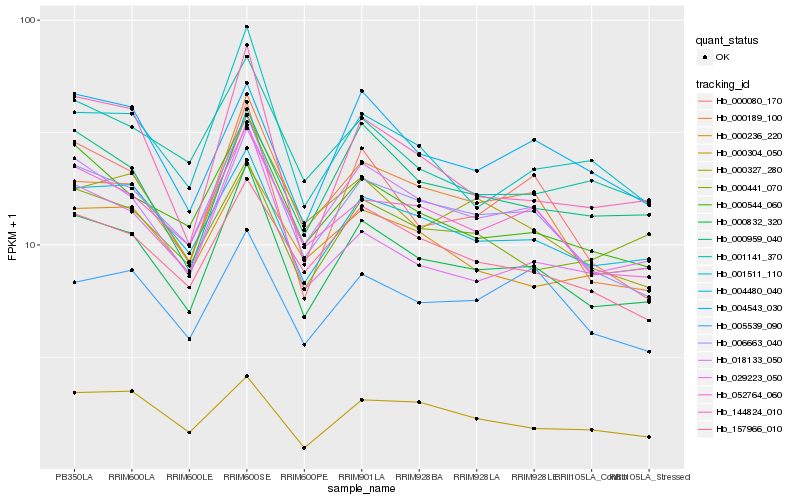
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_320 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_006663_040 |
0.0556237266 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_018133_050 |
0.0610273225 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 4 |
Hb_004480_040 |
0.0661219501 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_052764_060 |
0.0682104108 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000544_060 |
0.0751173948 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_000959_040 |
0.078757297 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 8 |
Hb_157966_010 |
0.0818783392 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 9 |
Hb_144824_010 |
0.0820849749 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 10 |
Hb_000189_100 |
0.0838511067 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 11 |
Hb_001511_110 |
0.0873920124 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000080_170 |
0.08777387 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000327_280 |
0.0883474142 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004543_030 |
0.090095738 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 15 |
Hb_000304_050 |
0.0902420285 |
transcription factor |
TF Family: GRAS |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 16 |
Hb_001141_370 |
0.0914861556 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 17 |
Hb_000441_070 |
0.0917356153 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000236_220 |
0.0941151769 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 19 |
Hb_029223_050 |
0.0949315629 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005539_090 |
0.0951553142 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |