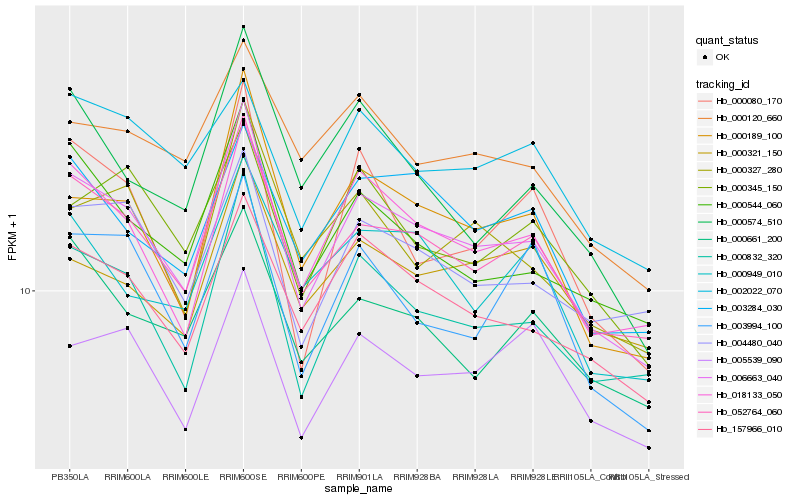
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006663_040 |
0.0 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_052764_060 |
0.0444042229 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000832_320 |
0.0556237266 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_018133_050 |
0.062385572 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 5 |
Hb_000189_100 |
0.0643647225 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 6 |
Hb_157966_010 |
0.0684363753 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 7 |
Hb_000544_060 |
0.0711874805 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 8 |
Hb_004480_040 |
0.0795657573 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000327_280 |
0.0802702775 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000661_200 |
0.082766989 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003284_030 |
0.0835043154 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_150 |
0.0867912198 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003994_100 |
0.0873235437 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000120_660 |
0.0890582696 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 15 |
Hb_005539_090 |
0.0892041027 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000574_510 |
0.0899155966 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000949_010 |
0.0904075691 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 18 |
Hb_000080_170 |
0.0921309145 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000321_150 |
0.0935092779 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002022_070 |
0.0944918635 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |