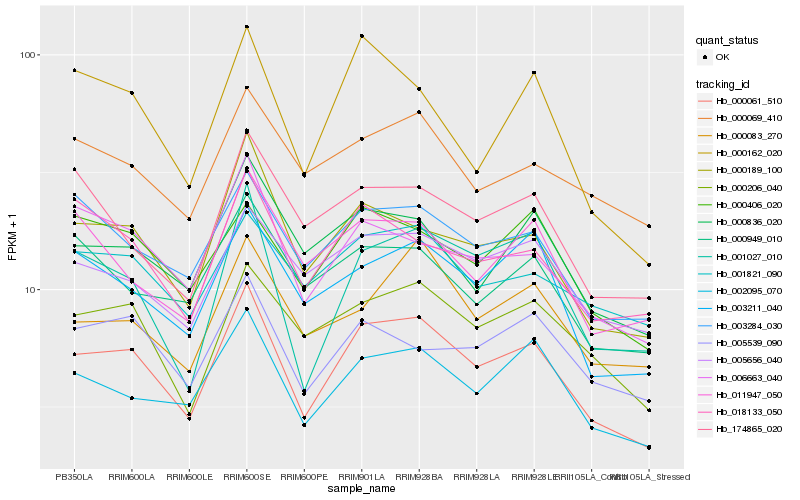
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_510 |
0.0 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 2 |
Hb_002095_070 |
0.0861307042 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 3 |
Hb_000189_100 |
0.0868366253 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 4 |
Hb_001027_010 |
0.0944528161 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0019s03720g [Populus trichocarpa] |
| 5 |
Hb_000206_040 |
0.0960113042 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 6 |
Hb_003211_040 |
0.09873335 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000836_020 |
0.1015641075 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 8 |
Hb_003284_030 |
0.1018892903 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006663_040 |
0.1022471679 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000162_020 |
0.1055593147 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 11 |
Hb_005656_040 |
0.1070763748 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_000406_020 |
0.1087504607 |
- |
- |
- |
| 13 |
Hb_018133_050 |
0.1091299805 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 14 |
Hb_005539_090 |
0.1095196701 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000083_270 |
0.1098380342 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001821_090 |
0.1108106965 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 17 |
Hb_000069_410 |
0.1124509967 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 18 |
Hb_011947_050 |
0.1137392735 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 19 |
Hb_174865_020 |
0.1138457929 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 20 |
Hb_000949_010 |
0.1144675193 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |