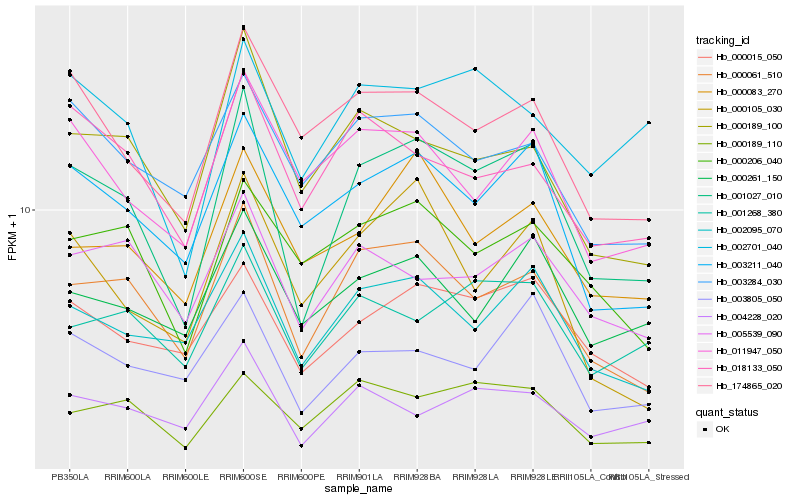
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001027_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0019s03720g [Populus trichocarpa] |
| 2 |
Hb_000061_510 |
0.0944528161 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 3 |
Hb_002095_070 |
0.1185985391 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 4 |
Hb_003211_040 |
0.1215828862 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000015_050 |
0.1268559783 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 6 |
Hb_174865_020 |
0.1335736959 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 7 |
Hb_000189_110 |
0.1338129332 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000189_100 |
0.134674134 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 9 |
Hb_004228_020 |
0.1360912421 |
- |
- |
hypothetical protein JCGZ_21866 [Jatropha curcas] |
| 10 |
Hb_003805_050 |
0.1366483594 |
- |
- |
- |
| 11 |
Hb_005539_090 |
0.137975336 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000083_270 |
0.1383699555 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001268_380 |
0.1386282023 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 14 |
Hb_000206_040 |
0.139477561 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 15 |
Hb_011947_050 |
0.1401460153 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 16 |
Hb_002701_040 |
0.1403993481 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 17 |
Hb_018133_050 |
0.1418842177 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 18 |
Hb_000261_150 |
0.1433797296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000105_030 |
0.1436489919 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 20 |
Hb_003284_030 |
0.1445876842 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |