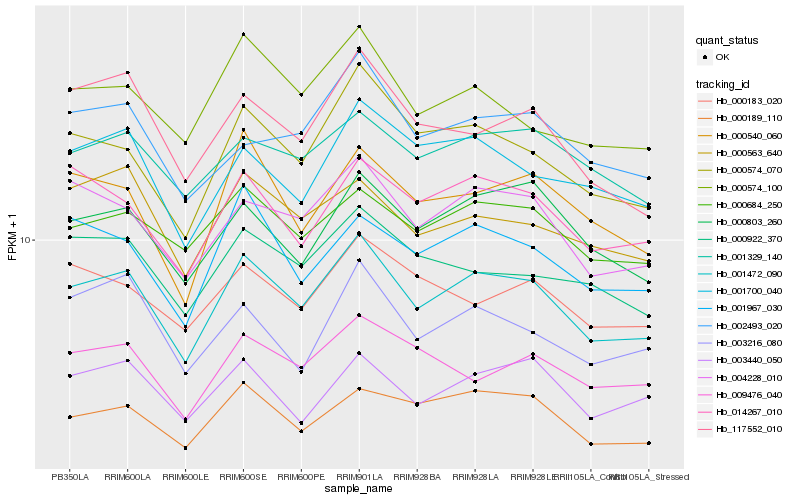
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_090 |
0.0 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004228_010 |
0.0690172025 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |
| 3 |
Hb_000574_070 |
0.0696553941 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 4 |
Hb_000803_260 |
0.079356829 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 5 |
Hb_014267_010 |
0.0795865441 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000684_250 |
0.0825493886 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 7 |
Hb_000540_060 |
0.0833399005 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 8 |
Hb_000574_100 |
0.0839251901 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 9 |
Hb_001700_040 |
0.0850913202 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 10 |
Hb_001967_030 |
0.0853407546 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 11 |
Hb_000922_370 |
0.0866971596 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 12 |
Hb_117552_010 |
0.0868455154 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000189_110 |
0.0881184913 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001329_140 |
0.0881987802 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 15 |
Hb_000563_640 |
0.0887433816 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 16 |
Hb_009476_040 |
0.0896549603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003440_050 |
0.0906182717 |
- |
- |
hypothetical protein JCGZ_05806 [Jatropha curcas] |
| 18 |
Hb_002493_020 |
0.0924192149 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000183_020 |
0.0927127954 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003216_080 |
0.0944198845 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560-like [Jatropha curcas] |