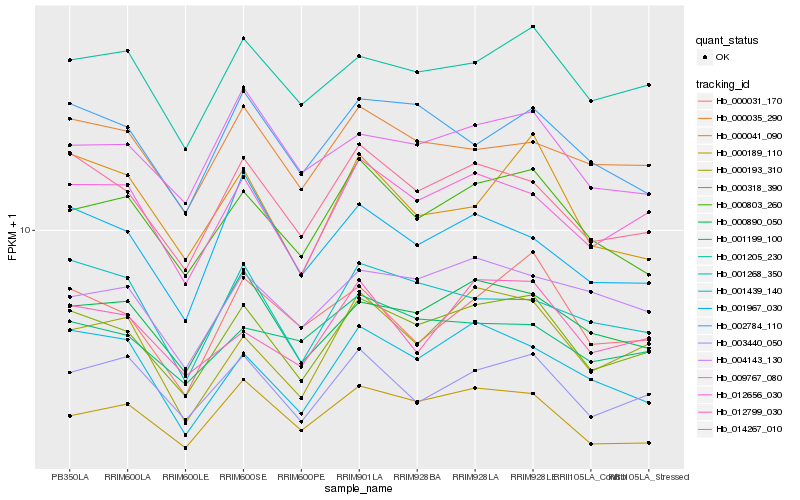
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631693 isoform X1 [Jatropha curcas] |
| 2 |
Hb_014267_010 |
0.0616896355 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012656_030 |
0.0617791299 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000890_050 |
0.0748245386 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 5 |
Hb_000803_260 |
0.0749200509 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 6 |
Hb_001205_230 |
0.077748127 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 7 |
Hb_003440_050 |
0.0827965869 |
- |
- |
hypothetical protein JCGZ_05806 [Jatropha curcas] |
| 8 |
Hb_002784_110 |
0.0890565893 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 9 |
Hb_000193_310 |
0.0891314408 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 10 |
Hb_004143_130 |
0.0900558001 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |
| 11 |
Hb_012799_030 |
0.0901624283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 12 |
Hb_009767_080 |
0.0907386594 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 13 |
Hb_000031_170 |
0.0917343935 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000035_290 |
0.09235962 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 15 |
Hb_000189_110 |
0.0933752414 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001439_140 |
0.0942128404 |
- |
- |
pattern formation protein, putative [Ricinus communis] |
| 17 |
Hb_001967_030 |
0.0942297639 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 18 |
Hb_001199_100 |
0.0942972502 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 19 |
Hb_000041_090 |
0.0943920019 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 20 |
Hb_001268_350 |
0.0946097976 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |