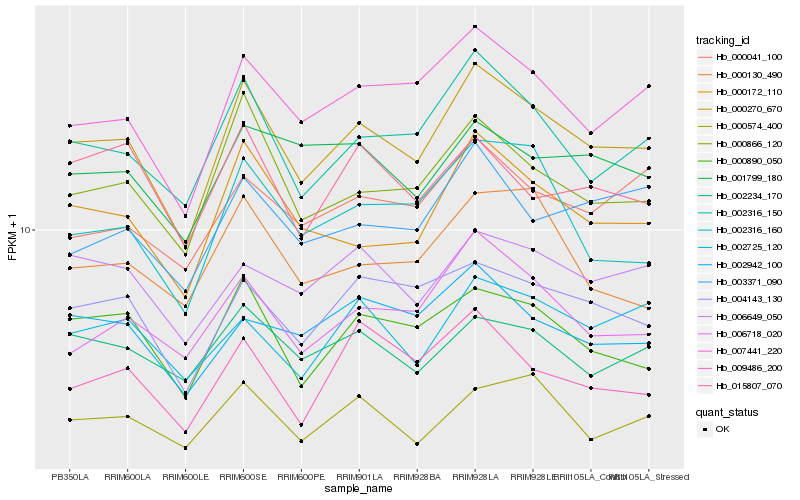
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_670 |
0.0 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 2 |
Hb_002316_150 |
0.0618735717 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 3 |
Hb_000172_110 |
0.064877021 |
- |
- |
- |
| 4 |
Hb_007441_220 |
0.066551999 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 5 |
Hb_000866_120 |
0.0670471855 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 6 |
Hb_000890_050 |
0.0712236707 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 7 |
Hb_004143_130 |
0.0737914855 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |
| 8 |
Hb_006718_020 |
0.0773861721 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000574_400 |
0.0826279795 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 10 |
Hb_006649_050 |
0.083160647 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 11 |
Hb_002234_170 |
0.0841351891 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002316_160 |
0.0852898017 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 13 |
Hb_003371_090 |
0.0859241155 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 14 |
Hb_000041_100 |
0.0871854432 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 15 |
Hb_002942_100 |
0.0871856361 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002725_120 |
0.0875669565 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001799_180 |
0.0892868168 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 18 |
Hb_000130_490 |
0.0903835872 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009486_200 |
0.0920866155 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 20 |
Hb_015807_070 |
0.0929620133 |
- |
- |
PREDICTED: autophagy-related protein 18f [Jatropha curcas] |