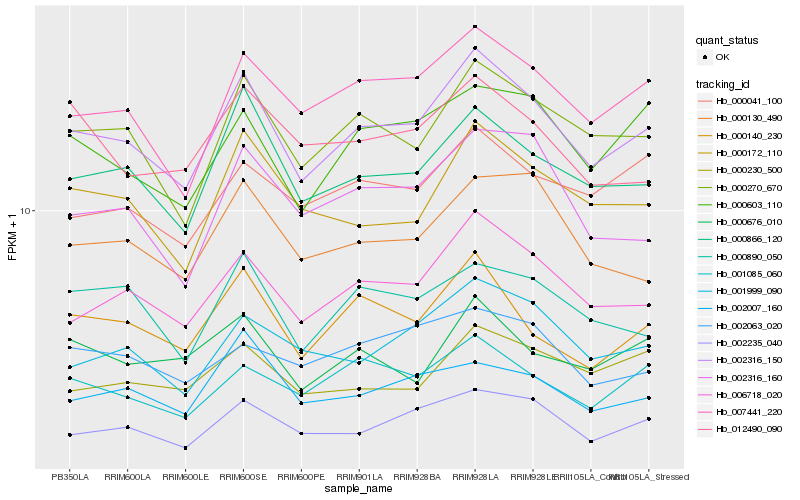
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002316_150 |
0.0 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 2 |
Hb_006718_020 |
0.0608381503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000270_670 |
0.0618735717 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 4 |
Hb_007441_220 |
0.0656860392 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 5 |
Hb_000866_120 |
0.0673539583 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 6 |
Hb_000172_110 |
0.0764227319 |
- |
- |
- |
| 7 |
Hb_000140_230 |
0.0771798907 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_002235_040 |
0.0792174898 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 9 |
Hb_000603_110 |
0.0803944048 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 10 |
Hb_001085_060 |
0.0824578906 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g04840 [Jatropha curcas] |
| 11 |
Hb_001999_090 |
0.0839746766 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002316_160 |
0.0858119844 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 13 |
Hb_000130_490 |
0.0862388078 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000890_050 |
0.0863085047 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 15 |
Hb_000041_100 |
0.0864125838 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 16 |
Hb_000230_500 |
0.0904452012 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000676_010 |
0.0919897564 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 18 |
Hb_002007_160 |
0.0923493191 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 19 |
Hb_002063_020 |
0.0937712243 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 20 |
Hb_012490_090 |
0.0945405717 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |