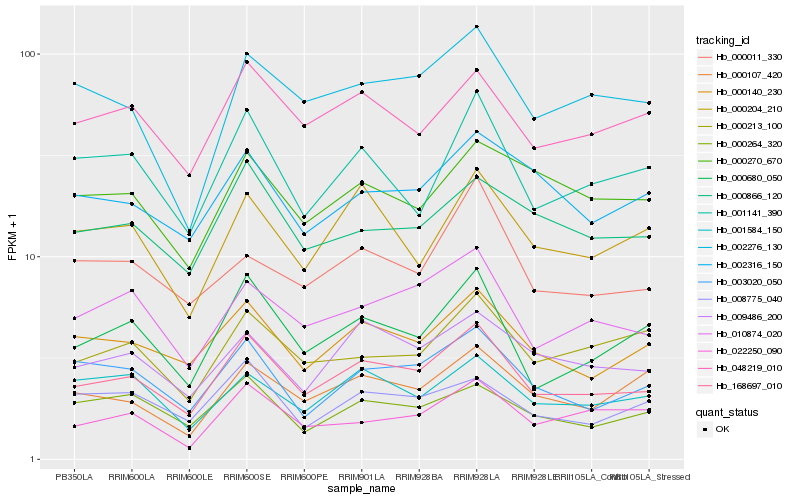
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168697_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 2 |
Hb_000140_230 |
0.0790427899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_000680_050 |
0.081682874 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 4 |
Hb_010874_020 |
0.0867710331 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 5 |
Hb_048219_010 |
0.0908423289 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 6 |
Hb_003020_050 |
0.0942421993 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_009486_200 |
0.0958383804 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 8 |
Hb_001141_390 |
0.097440966 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 9 |
Hb_000866_120 |
0.0997779548 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 10 |
Hb_002276_130 |
0.0999302095 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 11 |
Hb_000264_320 |
0.1025352748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 12 |
Hb_000204_210 |
0.1039038516 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 13 |
Hb_002316_150 |
0.107059871 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 14 |
Hb_008775_040 |
0.1073151694 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 15 |
Hb_001584_150 |
0.1082874022 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 16 |
Hb_000213_100 |
0.1095749967 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 17 |
Hb_000107_420 |
0.1096907141 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000011_330 |
0.1131016271 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 19 |
Hb_000270_670 |
0.1136547508 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 20 |
Hb_022250_090 |
0.1136822748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |