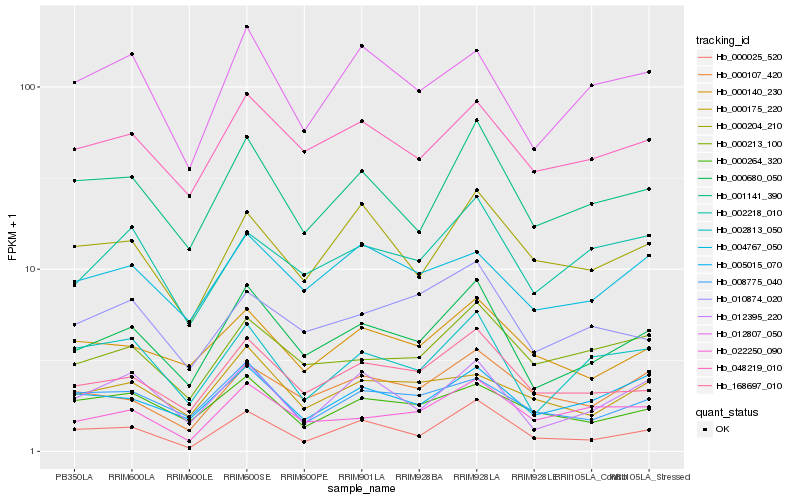
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 2 |
Hb_168697_010 |
0.081682874 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 3 |
Hb_001141_390 |
0.0900484651 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 4 |
Hb_048219_010 |
0.0968019679 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 5 |
Hb_012395_220 |
0.1042338994 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 6 |
Hb_002813_050 |
0.1075831445 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 7 |
Hb_005015_070 |
0.108398745 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 8 |
Hb_000213_100 |
0.1098758762 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 9 |
Hb_012807_050 |
0.1111418473 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 10 |
Hb_000140_230 |
0.1126903256 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000025_520 |
0.1153301363 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27110 [Jatropha curcas] |
| 12 |
Hb_002218_010 |
0.115443776 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 13 |
Hb_008775_040 |
0.1160082035 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 14 |
Hb_022250_090 |
0.1167256614 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 15 |
Hb_004767_050 |
0.1172034722 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 16 |
Hb_010874_020 |
0.1180070015 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 17 |
Hb_000204_210 |
0.1216042778 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 18 |
Hb_000107_420 |
0.1221760013 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000264_320 |
0.1223556772 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 20 |
Hb_000175_220 |
0.1236366897 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |