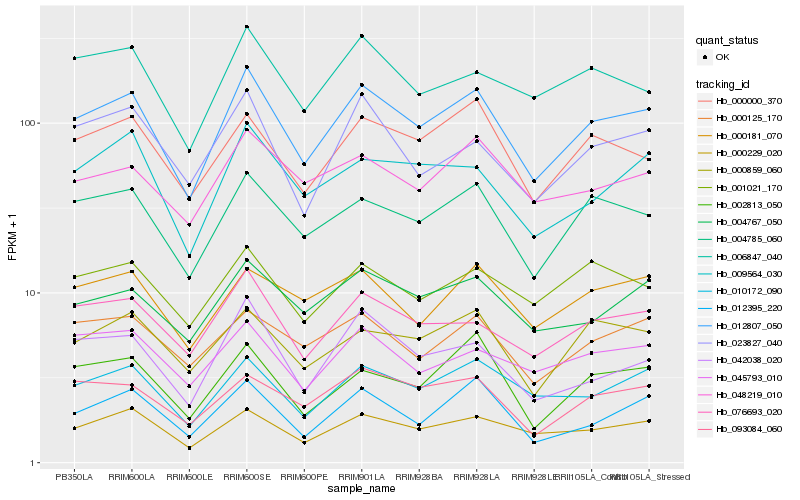
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012807_050 |
0.0 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 2 |
Hb_004785_060 |
0.0725652685 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 3 |
Hb_000229_020 |
0.0742071282 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03540 [Jatropha curcas] |
| 4 |
Hb_010172_090 |
0.0787030967 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_076693_020 |
0.0826368504 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 6 |
Hb_000000_370 |
0.0847859469 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 7 |
Hb_004767_050 |
0.0863535783 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 8 |
Hb_093084_060 |
0.0888877025 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 9 |
Hb_000859_060 |
0.0899937999 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 10 |
Hb_009564_030 |
0.0928889355 |
transcription factor |
TF Family: bHLH |
Transcription factor AtMYC2, putative [Ricinus communis] |
| 11 |
Hb_006847_040 |
0.0947784736 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 12 |
Hb_048219_010 |
0.0951877466 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 13 |
Hb_012395_220 |
0.0952939486 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 14 |
Hb_042038_020 |
0.0955868479 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 15 |
Hb_023827_040 |
0.0960908087 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 16 |
Hb_001021_170 |
0.0962845255 |
- |
- |
PREDICTED: uncharacterized protein LOC105642474 [Jatropha curcas] |
| 17 |
Hb_000125_170 |
0.096425137 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 18 |
Hb_045793_010 |
0.0964799386 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 19 |
Hb_000181_070 |
0.0966194409 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_002813_050 |
0.0968219728 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |