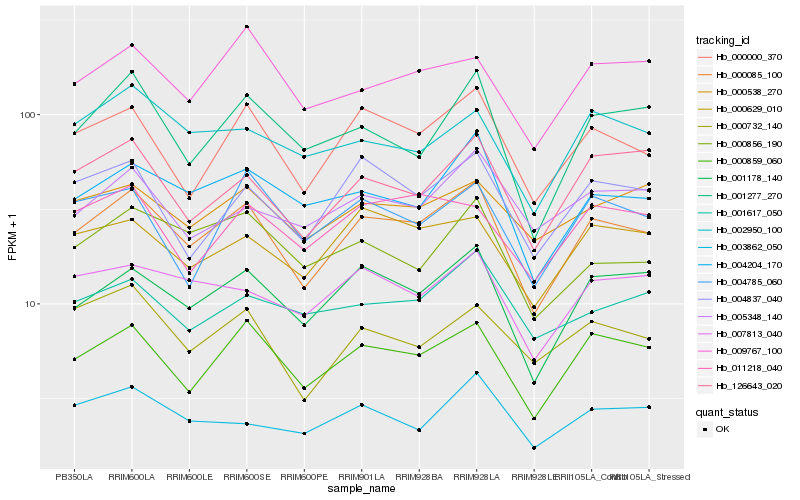
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_100 |
0.0 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 2 |
Hb_000859_060 |
0.0653672941 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 3 |
Hb_001178_140 |
0.0687673325 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000000_370 |
0.071707415 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 5 |
Hb_004204_170 |
0.084389951 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 6 |
Hb_126643_020 |
0.0900287175 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 7 |
Hb_000629_010 |
0.0900755558 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004837_040 |
0.0907677318 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 9 |
Hb_001277_270 |
0.0928047353 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007813_040 |
0.0936917107 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000856_190 |
0.0961314784 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 12 |
Hb_000732_140 |
0.0971852945 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC [Jatropha curcas] |
| 13 |
Hb_009767_100 |
0.0976750051 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 14 |
Hb_002950_100 |
0.1004950805 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 15 |
Hb_000538_270 |
0.1009768816 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 16 |
Hb_003862_050 |
0.1017781232 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 17 |
Hb_011218_040 |
0.1021603231 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_004785_060 |
0.1029807463 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 19 |
Hb_001617_050 |
0.1032728983 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005348_140 |
0.1041809486 |
- |
- |
PREDICTED: ubiquinone biosynthesis protein COQ9-B, mitochondrial [Jatropha curcas] |