| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
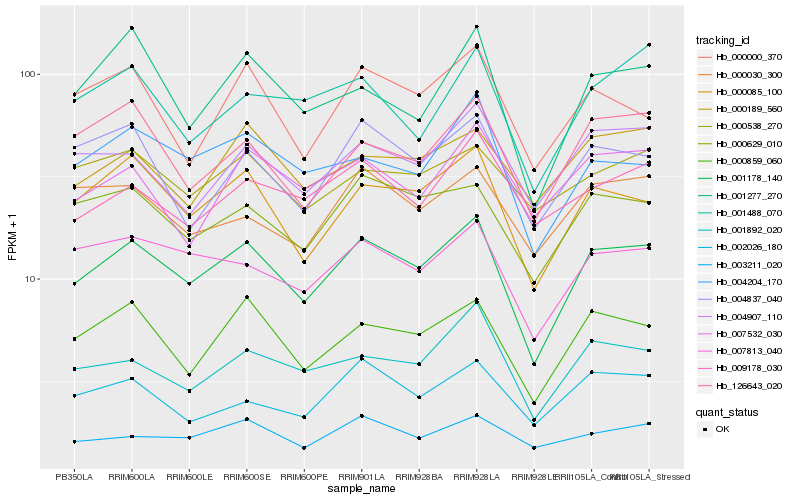
Hb_001178_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000085_100 |
0.0687673325 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 3 |
Hb_000859_060 |
0.0717514331 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 4 |
Hb_000629_010 |
0.0817399646 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007813_040 |
0.0821916955 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001892_020 |
0.0844728725 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_004204_170 |
0.0878968234 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 8 |
Hb_126643_020 |
0.0920908903 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 9 |
Hb_004907_110 |
0.0921423168 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001277_270 |
0.0926817195 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004837_040 |
0.0947324617 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 12 |
Hb_002026_180 |
0.095856071 |
- |
- |
PREDICTED: uncharacterized protein LOC105645805 [Jatropha curcas] |
| 13 |
Hb_000000_370 |
0.0974598712 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 14 |
Hb_003211_020 |
0.0981225343 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g08310, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000189_560 |
0.0981503934 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 16 |
Hb_000030_300 |
0.1004212643 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 17 |
Hb_000538_270 |
0.1015248024 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 18 |
Hb_009178_030 |
0.101620841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001488_070 |
0.1027523512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007532_030 |
0.102809574 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |