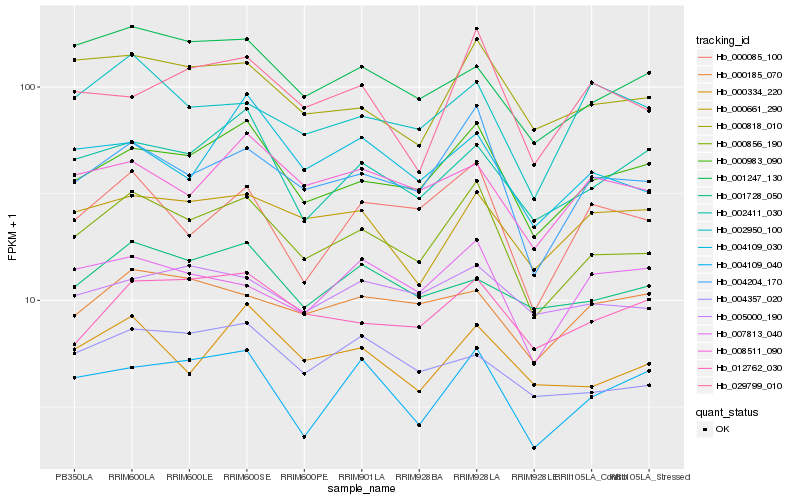
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_190 |
0.0 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 2 |
Hb_004204_170 |
0.0579943541 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 3 |
Hb_000983_090 |
0.0658863661 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 4 |
Hb_000818_010 |
0.0853349208 |
- |
- |
Calmodulin-domain protein kinase 5 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000334_220 |
0.0959951716 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 6 |
Hb_000085_100 |
0.0961314784 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 7 |
Hb_012762_030 |
0.0980577007 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 8 |
Hb_001247_130 |
0.0982961834 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 9 |
Hb_002411_030 |
0.0999844706 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 10 |
Hb_008511_090 |
0.1014645872 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 11 |
Hb_004109_040 |
0.1027070597 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007813_040 |
0.1035770083 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 13 |
Hb_029799_010 |
0.1037015431 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_000661_290 |
0.1037919701 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_004109_030 |
0.1040668768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004357_020 |
0.1055657936 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 17 |
Hb_002950_100 |
0.1061571573 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 18 |
Hb_001728_050 |
0.1067071344 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 19 |
Hb_005000_190 |
0.1073405967 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 20 |
Hb_000185_070 |
0.1079110514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |