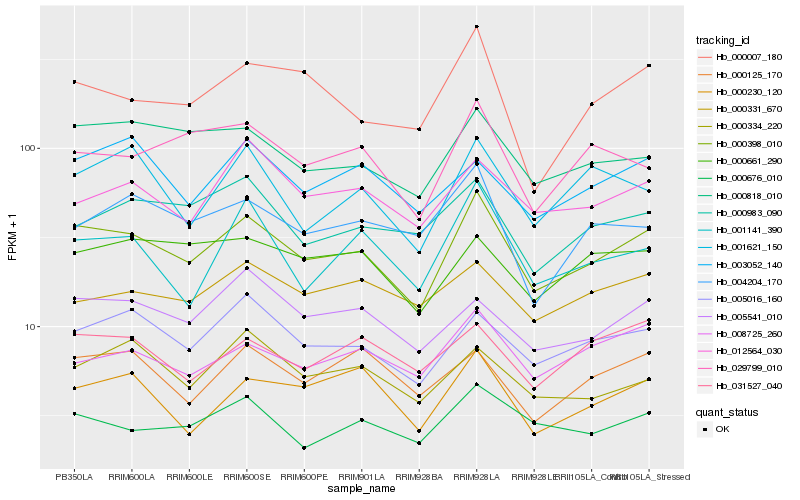
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005016_160 |
0.0887869477 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 3 |
Hb_000818_010 |
0.0918252246 |
- |
- |
Calmodulin-domain protein kinase 5 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000676_010 |
0.0982366774 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 5 |
Hb_005541_010 |
0.0995255186 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 6 |
Hb_008725_260 |
0.1025755513 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 7 |
Hb_000230_120 |
0.1034214601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 8 |
Hb_000007_180 |
0.1043442892 |
- |
- |
hypothetical protein PRUPE_ppb024585mg, partial [Prunus persica] |
| 9 |
Hb_029799_010 |
0.1048353392 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_001141_390 |
0.1056515073 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 11 |
Hb_000661_290 |
0.1065402609 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 12 |
Hb_012564_030 |
0.1065778937 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 13 |
Hb_000125_170 |
0.1085463924 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 14 |
Hb_031527_040 |
0.1088221882 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000983_090 |
0.1092485113 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 16 |
Hb_001621_150 |
0.1095218162 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 17 |
Hb_003052_140 |
0.1098115059 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 18 |
Hb_000334_220 |
0.1104627655 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 19 |
Hb_004204_170 |
0.1107025225 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 20 |
Hb_000331_670 |
0.1128204039 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |