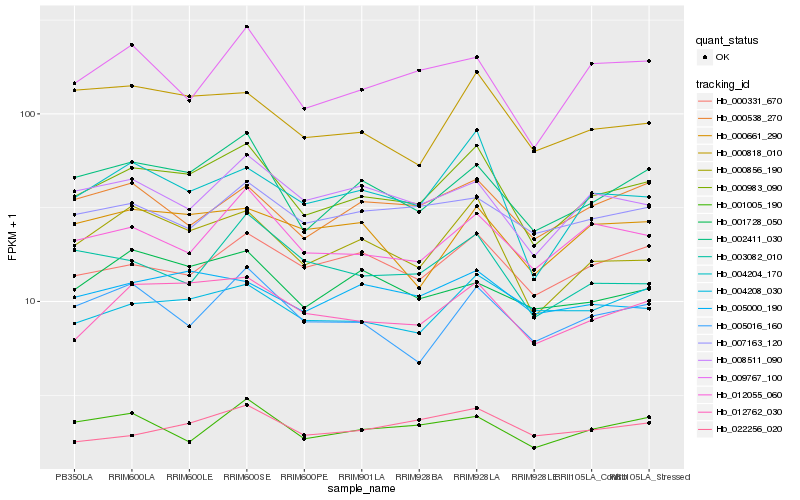
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000983_090 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 2 |
Hb_000856_190 |
0.0658863661 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 3 |
Hb_002411_030 |
0.0668639995 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 4 |
Hb_012762_030 |
0.0692317431 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 5 |
Hb_004204_170 |
0.0776080751 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 6 |
Hb_012055_060 |
0.0810047743 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000331_670 |
0.0862192414 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 8 |
Hb_008511_090 |
0.0882835012 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 9 |
Hb_005016_160 |
0.0899556293 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 10 |
Hb_009767_100 |
0.0910554337 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 11 |
Hb_000818_010 |
0.0915827252 |
- |
- |
Calmodulin-domain protein kinase 5 isoform 1 [Theobroma cacao] |
| 12 |
Hb_001005_190 |
0.0924053002 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004208_030 |
0.0947187835 |
transcription factor |
TF Family: MYB-related |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 14 |
Hb_000538_270 |
0.0953004503 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 15 |
Hb_000661_290 |
0.0973256224 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_003082_010 |
0.0984132288 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 17 |
Hb_005000_190 |
0.0984594308 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 18 |
Hb_007163_120 |
0.0988742954 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 19 |
Hb_022256_020 |
0.0993420411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35130, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001728_050 |
0.1001794703 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |