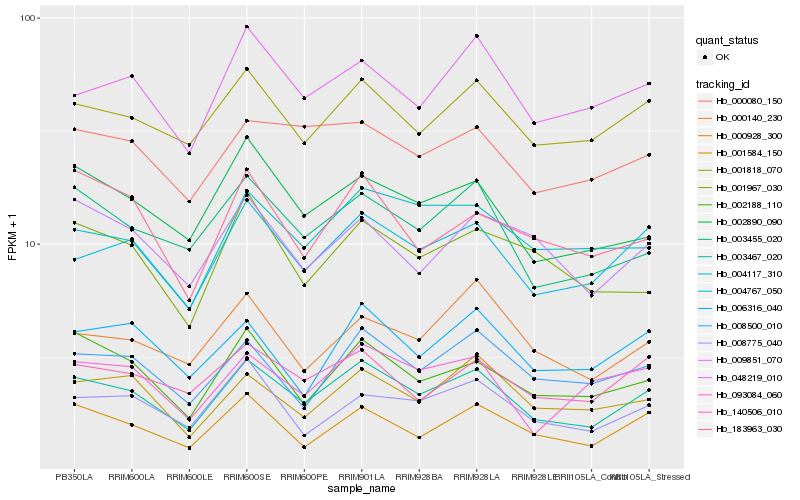
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003467_020 |
0.0 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 13-like [Jatropha curcas] |
| 2 |
Hb_004767_050 |
0.0749502436 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 3 |
Hb_003455_020 |
0.0845498728 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 4 |
Hb_002188_110 |
0.0873251242 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_002890_090 |
0.0886596612 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 6 |
Hb_001818_070 |
0.0886657561 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 7 |
Hb_008500_010 |
0.0915843415 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 8 |
Hb_140506_010 |
0.0928897474 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170-like [Jatropha curcas] |
| 9 |
Hb_048219_010 |
0.0933973154 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 10 |
Hb_000928_300 |
0.0941221271 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g03580 [Jatropha curcas] |
| 11 |
Hb_001584_150 |
0.0954496867 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 12 |
Hb_004117_310 |
0.0981215568 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 13 |
Hb_000080_150 |
0.0994599768 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006316_040 |
0.1001398082 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 15 |
Hb_008775_040 |
0.1004462754 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 16 |
Hb_183963_030 |
0.1008053094 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000140_230 |
0.101076363 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_093084_060 |
0.101686359 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 19 |
Hb_009851_070 |
0.102467994 |
- |
- |
PREDICTED: cyclin-dependent kinase E-1 [Jatropha curcas] |
| 20 |
Hb_001967_030 |
0.1030202077 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |