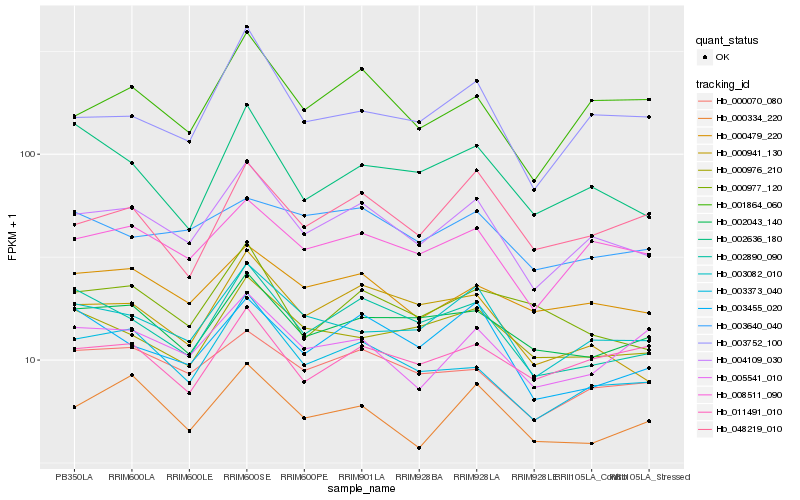
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008511_090 |
0.0564954549 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 3 |
Hb_003082_010 |
0.0652123683 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 4 |
Hb_003373_040 |
0.0683996155 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 5 |
Hb_002890_090 |
0.0712533008 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 6 |
Hb_000070_080 |
0.0719825793 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000941_130 |
0.0727158318 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 8 |
Hb_000479_220 |
0.0730475111 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 9 |
Hb_001864_060 |
0.0735248125 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 10 |
Hb_002043_140 |
0.0789563479 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 11 |
Hb_003752_100 |
0.0793620563 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 12 |
Hb_000334_220 |
0.0802572852 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 13 |
Hb_003455_020 |
0.0847248917 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 14 |
Hb_000976_210 |
0.0870749713 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005541_010 |
0.0881138994 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 16 |
Hb_048219_010 |
0.0898037693 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 17 |
Hb_011491_010 |
0.0901784018 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 18 |
Hb_003640_040 |
0.09108947 |
- |
- |
PREDICTED: uncharacterized protein LOC105647348 [Jatropha curcas] |
| 19 |
Hb_000977_120 |
0.092735982 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002636_180 |
0.0932743315 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |