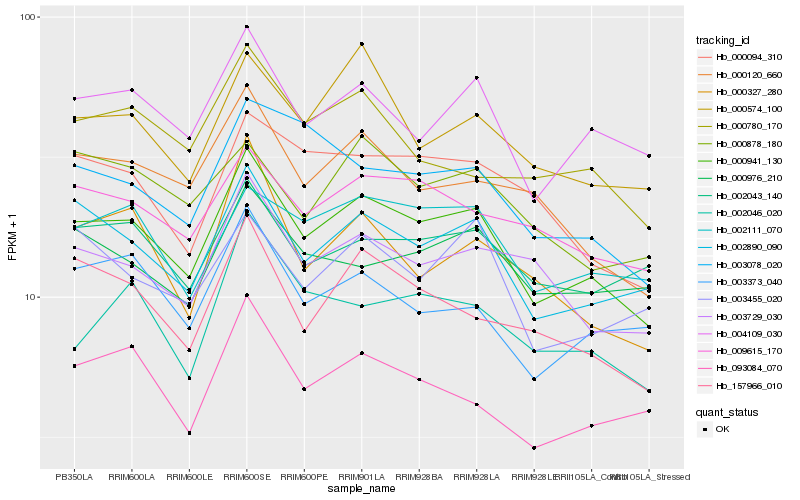
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000941_130 |
0.0 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 2 |
Hb_002890_090 |
0.0657640618 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 3 |
Hb_004109_030 |
0.0727158318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002111_070 |
0.0779083416 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 5 |
Hb_009615_170 |
0.0788702344 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_157966_010 |
0.0815710208 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 7 |
Hb_003729_030 |
0.0828609263 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 8 |
Hb_000120_660 |
0.0844250884 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 9 |
Hb_000574_100 |
0.0845678493 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 10 |
Hb_003078_020 |
0.0866738559 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 11 |
Hb_000094_310 |
0.0872375417 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 12 |
Hb_002043_140 |
0.0886778047 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 13 |
Hb_003373_040 |
0.0887227018 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 14 |
Hb_093084_070 |
0.0894185483 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 15 |
Hb_000878_180 |
0.0910148591 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000976_210 |
0.0918326657 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003455_020 |
0.0923049234 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 18 |
Hb_002046_020 |
0.0930608046 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 19 |
Hb_000780_170 |
0.0934495008 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 20 |
Hb_000327_280 |
0.0938768222 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |