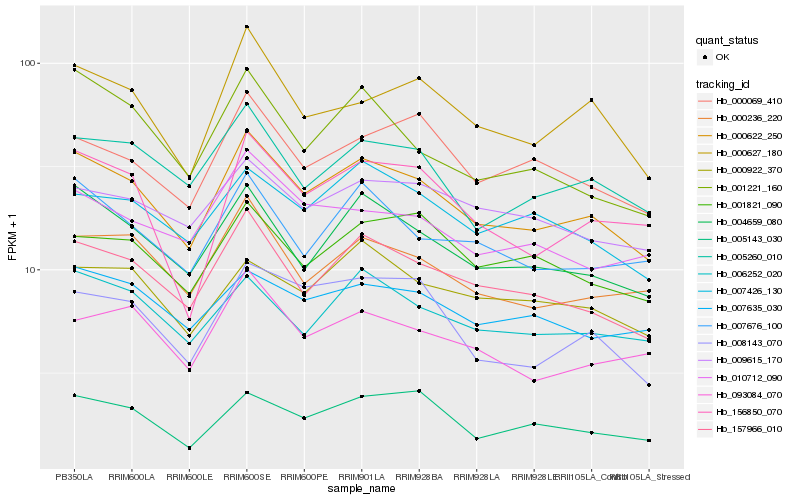
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_250 |
0.0 |
- |
- |
atpob1, putative [Ricinus communis] |
| 2 |
Hb_157966_010 |
0.0567716892 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 3 |
Hb_004659_080 |
0.0645830863 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 4 |
Hb_000236_220 |
0.0750499316 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 5 |
Hb_005260_010 |
0.0768920947 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000627_180 |
0.0776542754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006252_020 |
0.0801433821 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007676_100 |
0.0804787413 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 9 |
Hb_005143_030 |
0.0822901951 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 10 |
Hb_007426_130 |
0.0838753031 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_156850_070 |
0.0843479679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008143_070 |
0.0856893097 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 13 |
Hb_007635_030 |
0.0863693112 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000069_410 |
0.0872936152 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 15 |
Hb_001821_090 |
0.087621136 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 16 |
Hb_010712_090 |
0.0884303651 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 17 |
Hb_000922_370 |
0.0885946855 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 18 |
Hb_093084_070 |
0.0888251293 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_009615_170 |
0.0889248364 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001221_160 |
0.089455216 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |