| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
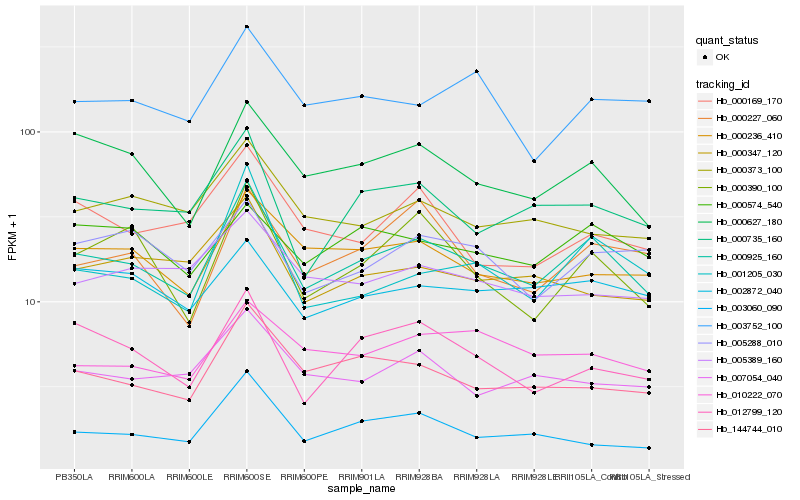
Hb_000925_160 |
0.0 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000627_180 |
0.097840025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000735_160 |
0.1037428598 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 4 |
Hb_144744_010 |
0.1073198754 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 5 |
Hb_000236_410 |
0.1101607881 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 6 |
Hb_001205_030 |
0.1140415688 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 7 |
Hb_000390_100 |
0.116124933 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 8 |
Hb_007054_040 |
0.11694204 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 9 |
Hb_000227_060 |
0.1194392235 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 10 |
Hb_012799_120 |
0.1199430942 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002872_040 |
0.1204283417 |
- |
- |
pumilio, putative [Ricinus communis] |
| 12 |
Hb_010222_070 |
0.1213840959 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_003752_100 |
0.1216275725 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 14 |
Hb_000373_100 |
0.1216680495 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 15 |
Hb_000169_170 |
0.1235140714 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 16 |
Hb_005288_010 |
0.1248897817 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000347_120 |
0.1254549417 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 18 |
Hb_005389_160 |
0.1256107577 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 19 |
Hb_000574_540 |
0.1261659933 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_003060_090 |
0.126309777 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |