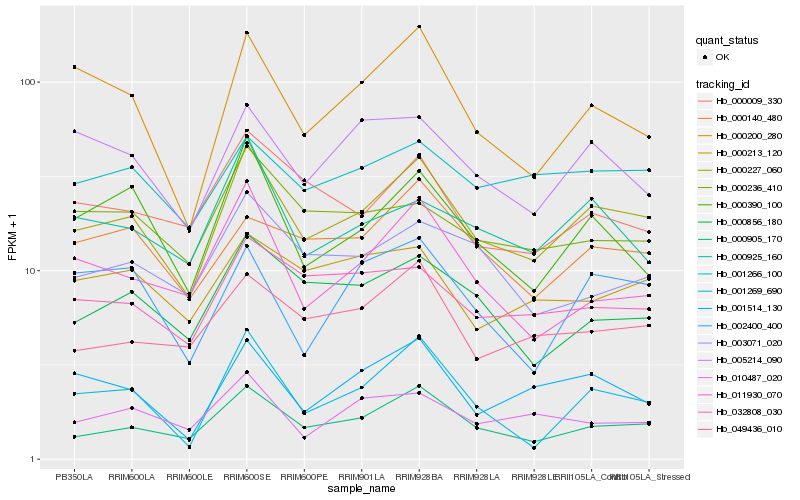
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_060 |
0.0 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 2 |
Hb_000905_170 |
0.0802545101 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 3 |
Hb_000200_280 |
0.1132840703 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 4 |
Hb_000390_100 |
0.1141407342 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 5 |
Hb_049436_010 |
0.1156720098 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 6 |
Hb_001514_130 |
0.1169735085 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 7 |
Hb_000856_180 |
0.1177158612 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 8 |
Hb_032808_030 |
0.1182275471 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000925_160 |
0.1194392235 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001266_100 |
0.1203567008 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 11 |
Hb_005214_090 |
0.1206752111 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 12 |
Hb_000009_330 |
0.1233648051 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_000140_480 |
0.1239890385 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 14 |
Hb_003071_020 |
0.1257102957 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 15 |
Hb_011930_070 |
0.1258960934 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 16 |
Hb_000236_410 |
0.1268649094 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 17 |
Hb_010487_020 |
0.1274873692 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 18 |
Hb_002400_400 |
0.1292259354 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_000213_120 |
0.129583627 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_001269_690 |
0.1326601752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |