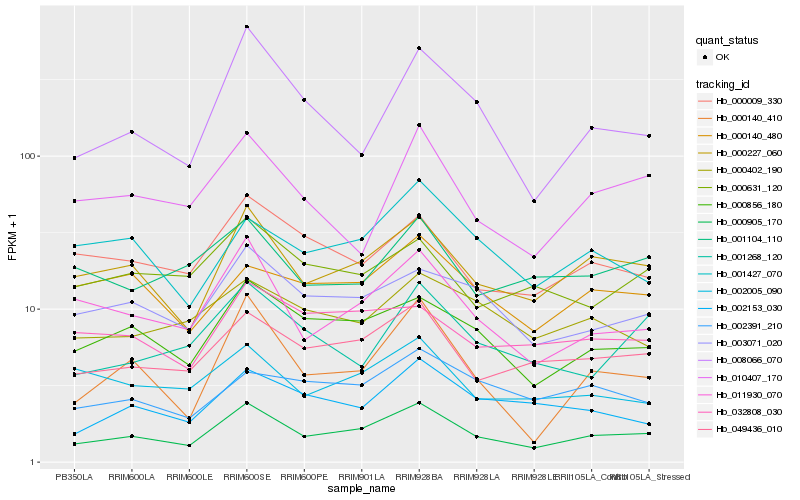
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_170 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 2 |
Hb_000227_060 |
0.0802545101 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 3 |
Hb_049436_010 |
0.0985676731 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 4 |
Hb_000856_180 |
0.1019408865 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 5 |
Hb_011930_070 |
0.1114983237 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 6 |
Hb_003071_020 |
0.1144972175 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 7 |
Hb_000009_330 |
0.1302530969 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000140_410 |
0.1309550816 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002153_030 |
0.134791576 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 10 |
Hb_000140_480 |
0.1356468645 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 11 |
Hb_001427_070 |
0.1358337646 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 12 |
Hb_008066_070 |
0.1382211154 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 13 |
Hb_002005_090 |
0.1384550956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010407_170 |
0.1385776078 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 15 |
Hb_002391_210 |
0.1389212219 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 16 |
Hb_001268_120 |
0.1403566647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_032808_030 |
0.1407548138 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000402_190 |
0.141184443 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 19 |
Hb_001104_110 |
0.1422203853 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 20 |
Hb_000631_120 |
0.142766396 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |